Streptococcus infantis, Streptococcus mitis, and Streptococcus oralis Strains With Highly Similar cps5 Loci and Antigenic Relatedness to Serotype 5 Pneumococci
- PMID: 30671034
- PMCID: PMC6332807
- DOI: 10.3389/fmicb.2018.03199
Streptococcus infantis, Streptococcus mitis, and Streptococcus oralis Strains With Highly Similar cps5 Loci and Antigenic Relatedness to Serotype 5 Pneumococci
Abstract
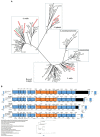
Streptococcus pneumoniae is a highly impactful bacterial pathogen on a global scale. The principal pneumococcal virulence factor and target of effective vaccines is its polysaccharide capsule, of which there are many structurally distinct forms. Here, we describe four distinct strains of three Mitis group commensal species (Streptococcus infantis, Streptococcus mitis, and Streptococcus oralis) recovered from upper respiratory tract specimens from adults in Kenya and the United States that were PCR-positive for the pneumococcal serotype 5 specific gene, wzy5. For each of the four strains, the 15 genes comprising the capsular polysaccharide biosynthetic gene cluster (cps5) shared the same order found in serotype 5 pneumococci, and each of the serotype 5-specific genes from the serotype 5 pneumococcal reference strain shared 76-99% sequence identity with the non-pneumococcal counterparts. Double-diffusion experiments demonstrated specific reactivity of the non-pneumococcal strains with pneumococcal serotype 5 typing sera. Antiserum raised against S. mitis strain KE67013 specifically reacted with serotype 5 pneumococci for a positive Quellung reaction and stimulated serotype 5 specific opsonophagocytic killing of pneumococci. Four additional commensal strains, identified using PCR serotyping assays on pharyngeal specimens, revealed loci highly homologous to those of pneumococci of serotypes 12F, 15A, 18C, and 33F. These data, in particular the species and strain diversity shown for serotype 5, highlight the existence of a broad non-pneumococcal species reservoir in the upper respiratory tract for the expression of capsular polysaccharides that are structurally related or identical to those corresponding to epidemiologically significant serotypes. Very little is known about the genetic and antigenic capsular diversity among the vast array of commensal streptococcal strains that represent multiple diverse species. The discovery of serotype 5 strains within three different commensal species suggests that extensive capsular serologic overlap exists between pneumococci and other members of the diverse Mitis group. These findings may have implications for our current understanding of naturally acquired immunity to S. pneumoniae and pneumococcal serotype distributions in different global regions. Further characterization of commensal strains carrying homologs of serotype-specific genes previously thought to be specific for pneumococci of known serotypes may shed light on the evolution of these important loci.
Keywords: capsular biosynthetic; capsular serotype; immunodiffusion analysis; opsonophagocytic killing (OPK); serotype-specific gene.
Figures

References
-
- Balicer R. D., Zarka S., Levine H., Klement E., Sela T., Porat N., et al. (2010). Control of Streptococcus pneumoniae serotype 5 epidemic of severe pneumonia among young army recruits by mass antibiotic treatment and vaccination. Vaccine 28 5591–5596. 10.1016/j.vaccine.2010.06.031 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

