Dissemination of CTX-M-Producing Escherichia coli in Freshwater Fishes From a French Watershed (Burgundy)
- PMID: 30671043
- PMCID: PMC6331413
- DOI: 10.3389/fmicb.2018.03239
Dissemination of CTX-M-Producing Escherichia coli in Freshwater Fishes From a French Watershed (Burgundy)
Abstract
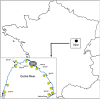
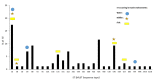
The burden of extended-spectrum β-lactamases producing Escherichia coli (ESBL-Ec), has increased over several decades. Freshwater ecosystems are suspected to play an important ecological and evolutionary role in driving the dissemination of antimicrobial resistance. The aim of our study was to decipher the occurrence of ESBL-Ec in a small watershed (Ouche river, Burgundy, France), targeting environmental matrices and fishes. Among cefotaxime resistant E. coli (ctxR Ec) isolates, we detected and characterized 36 ESBL-Ec from water, biofilm and fish guts. ctxR Ec and ESBL-Ec were found in samples from sites near the first small town, located downstream from the watershed which was studied. Treatment of urban wastewater by waste water treatment plants (WWTP), might therefore be a major potential source of ctxR Ec and thus of ESBL-Ec. Prevalence of total E. coli and ctxR Ec in fish guts ranged between 0 to 92% and 0 to 85%; respectively, depending on the sampling site and the fish species. The diet of fish (predator or omnivore) seems to strongly influence the prevalence of total E. coli and ESBL-Ec. Extended spectrum beta-lactamases produced by the isolates from this study belonged to the CTX-M family (CTX-M group 1 and 9). Moreover, some environmental ESBL-Ec proved to share genotypic features (MLST types) with isolates which originated from 8 WWTP effluents discharged in the Ouche river and with the sequence type ST131, which is widely described in clinical isolates. Ninety-seven % (97%) of ESBL-Ec from the study harbored additional antibiotic resistances and can thus be considered as multi drug resistant (MDR) bacteria. Finally, 53% of the ESBL-Ec strains harbored class 1 integron-integrase (intl1). These results are discussed with the perspective of defining indicators of antibiotic resistance contamination in freshwater ecosystems.
Keywords: ESBL producing Escherichia coli; MLST E. coli; antibiotic resistance; blaCTX–M; class 1 integron-integrase; fish; freshwater.
Figures



References
-
- Abgottspon H., Nuesch-Inderbinen M. T., Zurfluh K., Althaus D., Hachler H., Stephan R. (2014). Enterobacteriaceae with extended-spectrum- and pAmpC-type beta-lactamase-encoding genes isolated from freshwater fish from two lakes in Switzerland. Antimicrob. Agents Chemother. 58 2482–2484. 10.1128/aac.02689-13 - DOI - PMC - PubMed
-
- Blaak H., van Hoek A. H. A. M., Veenman C., van Leeuwen A. E. D., Lynch G., van Overbeek W. M., et al. (2014). Extended spectrum beta-lactamase- and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment. Int. J. Food Microbiol. 168 8–16. 10.1016/j.ijfoodmicro.2013.10.006 - DOI - PubMed
LinkOut - more resources
Full Text Sources

