Pregnancy Epigenetic Signature in T Helper 17 and T Regulatory Cells in Multiple Sclerosis
- PMID: 30671056
- PMCID: PMC6331474
- DOI: 10.3389/fimmu.2018.03075
Pregnancy Epigenetic Signature in T Helper 17 and T Regulatory Cells in Multiple Sclerosis
Abstract
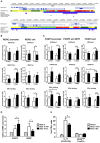
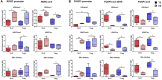
Increasing evidence supports the anti-inflammatory role of estrogens in Multiple Sclerosis (MS), originating from the observation of reduction in relapse rates among women with MS during pregnancy, but the molecular mechanisms are still not completely understood. Using an integrative data analysis, we identified T helper (Th) 17 and T regulatory (Treg) cell-type-specific regulatory regions (CSR) regulated by estrogen receptor alpha (ERα). These CSRs were validated in polarized Th17 from healthy donors (HD) and in peripheral blood mononuclear cells, Th17 and Treg cells from relapsing remitting (RR) MS patients and HD during pregnancy. 17β-estradiol induces active histone marks enrichment at Forkhead Box P3 (FOXP3)-CSRs and repressive histone marks enrichment at RAR related orphan receptor C (RORC)-CSRs in polarized Th17 cells. A disease-associated epigenetic profile was found in RRMS patients during pregnancy, suggesting a FOXP3 positive regulation and a RORC negative regulation in the third trimester of pregnancy. Altogether, these data indicate that estrogens act as immunomodulatory factors on the epigenomes of CD4+ T cells in RRMS; the identified CSRs may represent potential biomarkers for monitoring disease progression or new potential therapeutic targets.
Keywords: ERα; FOXP3; RORC; Th17; Treg; epigenetic profile; multiple sclerosis; pregnancy.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

