Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children
- PMID: 30671194
- PMCID: PMC6327923
- DOI: 10.1080/20002297.2018.1557986
Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children
Abstract
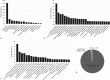
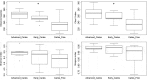
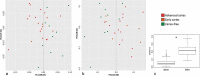
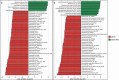
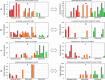
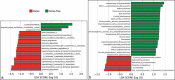
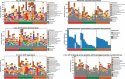
Studies of the microbiome associated with dental caries have largely relied on 16S rRNA sequence analysis, which is associated with PCR biases, low taxonomic resolution, and inability to accurately study functions. Here, we employed whole metagenome shotgun sequencing, coupled with high-resolution analysis algorithm, to analyze supragingival microbiomes from 30 children with or without dental caries. A total of 726 bacterial strains belonging to 406 species, in addition to 34 bacteriophages were identified. A core bacteriome was identified at the species and strain levels. Species of Prevotella, Veillonella, as yet unnamed Actinomyces, and Atopobium showed strongest association with caries; Streptococcus sp. AS14 and Leptotrichia sp. Oral taxon 225, among others, were overabundant in caries-free. For several species, the association was strain-specific. Furthermore, for some species, e.g. Streptococcus mitis and Streptococcus sanguinis, sister strains showed differential associations. Noteworthy, associations were also identified for phages: Streptococcus phage M102 with caries and Haemophilus phage HP1 with caries-free. Functionally, potentially relevant features were identified including urate, vitamin K2, and polyamine biosynthesis in association with caries; and three deiminases and lactate dehydrogenase with health. The results demonstrate new associations between the microbiome and dental caries at the strain and functional levels that need further investigation.
Keywords: Bacteria; High-throughput nucleotide sequencing; dental caries; metagenome; microbiota.
Figures
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
