Extremely low levels of chloroplast genome sequence variability in Astelia pumila (Asteliaceae, Asparagales)
- PMID: 30671303
- PMCID: PMC6339776
- DOI: 10.7717/peerj.6244
Extremely low levels of chloroplast genome sequence variability in Astelia pumila (Asteliaceae, Asparagales)
Abstract
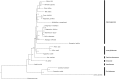
Astelia pumila (G.Forst.) Gaudich. (Asteliaceae, Asparagales) is a major element of West Patagonian cushion peat bog vegetation. With the aim to identify appropriate chloroplast markers for the use in a phylogeographic study, the complete chloroplast genomes of five A. pumila accessions from almost the entire geographical range of the species were assembled and screened for variable positions. The chloroplast genome sequence was obtained via a mapping approach, using Eustrephus latifolius (Asparagaceae) as a reference. The chloroplast genome of A. pumila varies in length from 158,215 bp to 158,221 bp, containing a large single copy region of 85,981-85,983 bp, a small single copy region of 18,182-18,186 bp and two inverted repeats of 27,026 bp. Genome annotation predicted a total of 113 genes, including 30 tRNA and four rRNA genes. Sequence comparisons revealed a very low degree of intraspecific genetic variability, as only 37 variable sites (18 indels, 18 single nucleotide polymorphisms, one 3-bp mutation)-most of them autapomorphies-were found among the five assembled chloroplast genomes. A Maximum Likelihood analysis, based on whole chloroplast genome sequences of several Asparagales accessions representing six of the currently recognized 14 families (sensu APG IV), confirmed the phylogenetic position of A. pumila. The chloroplast genome of A. pumila is the first to be reported for a member of the astelioid clade (14 genera with c. 215 species), a basally branching group within Asparagales.
Keywords: Asteliaceae; Chloroplast; Cushion plants; Genetic variability; Magellanic moorland; Organellar genome; Plastome; South America.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

References
-
- APG IV An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG I.V. Botanical Journal of the Linnean Society. 2016;181:1–20. doi: 10.1111/boj.12385. - DOI
-
- Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Molecular Biology and Evolution. 2014;31(12):3095–3112. doi: 10.1093/molbev/msu252. - DOI - PubMed
-
- Bayer C, Appel O, Rudall PJ. Asteliaceae. In: Kubitzki K, Huber H, editors. Flowering plants, monocotyledons: Lilianae (except Orchidaceae) Springer; Berlin: 1998. pp. 141–145.
LinkOut - more resources
Full Text Sources

