The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice
- PMID: 30671680
- PMCID: PMC6342742
- DOI: 10.1186/s12284-018-0260-z
The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice
Abstract
Background: Seed dormancy directly affects the phenotype of pre-harvest sprouting, and ultimately affects the quality and yield of rice seeds. Although many genes controlling seed dormancy have been cloned from cereals, the regulatory mechanisms controlling this process are complex, and much remains unknown. The MAPK cascade is involved in many signal transduction pathways. Recently, MKK3 has been reported to be involved in the regulation of seed dormancy, but its mechanism of action is unclear.
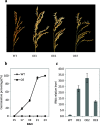
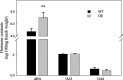
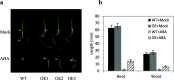
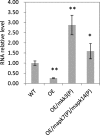
Results: We found that MKKK62-overexpressing rice lines (OE) lost seed dormancy. Further analyses showed that the abscisic acid (ABA) sensitivity of OE lines was decreased. In yeast two-hybrid experiments, MKKK62 interacted with MKK3, and MKK3 interacted with MAPK7 and MAPK14. Knock-out experiments confirmed that MKK3, MAPK7, and MAPK14 were involved in the regulation of seed dormancy. The OE lines showed decreased transcript levels of OsMFT, a homolog of a gene that controls seed dormancy in wheat. The up-regulation of OsMFT in MKK3-knockout lines (OE/mkk3) and MAPK7/14-knockout lines (OE/mapk7/mapk14) indicated that the MKKK62-MKK3-MAPK7/MAPK14 system controlled seed dormancy by regulating the transcription of OsMFT.
Conclusion: Our results showed that MKKK62 negatively controls seed dormancy in rice, and that during the germination stage and the late stage of seed maturation, ABA sensitivity and OsMFT transcription are negatively controlled by MKKK62. Our results have clarified the entire MAPK cascade controlling seed dormancy in rice. Together, these results indicate that protein modification by phosphorylation plays a key role in controlling seed dormancy.
Keywords: Dormancy; MAPK cascade; Pre-harvest sprouting; Rice (Oryza sativa L.).
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
MKK3 Cascade Regulates Seed Dormancy Through a Negative Feedback Loop Modulating ABA Signal in Rice.Rice (N Y). 2024 Jan 3;17(1):2. doi: 10.1186/s12284-023-00679-4. Rice (N Y). 2024. PMID: 38170405 Free PMC article.
-
The MKK3 module integrates nitrate and light signals to modulate secondary dormancy in Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2403646121. doi: 10.1073/pnas.2403646121. Epub 2024 Sep 19. Proc Natl Acad Sci U S A. 2024. PMID: 39298469 Free PMC article.
-
Current Insights into Weak Seed Dormancy and Pre-Harvest Sprouting in Crop Species.Plants (Basel). 2024 Sep 12;13(18):2559. doi: 10.3390/plants13182559. Plants (Basel). 2024. PMID: 39339534 Free PMC article. Review.
-
Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison.Funct Integr Genomics. 2004 May;4(2):84-93. doi: 10.1007/s10142-004-0104-3. Epub 2004 Feb 10. Funct Integr Genomics. 2004. PMID: 14770301
-
Seed Dormancy and Pre-Harvest Sprouting in Rice-An Updated Overview.Int J Mol Sci. 2021 Oct 30;22(21):11804. doi: 10.3390/ijms222111804. Int J Mol Sci. 2021. PMID: 34769234 Free PMC article. Review.
Cited by
-
MKK3 Cascade Regulates Seed Dormancy Through a Negative Feedback Loop Modulating ABA Signal in Rice.Rice (N Y). 2024 Jan 3;17(1):2. doi: 10.1186/s12284-023-00679-4. Rice (N Y). 2024. PMID: 38170405 Free PMC article.
-
The MKK3 MAPK cascade integrates temperature and after-ripening signals to modulate seed germination.Proc Natl Acad Sci U S A. 2024 Jul 9;121(28):e2404887121. doi: 10.1073/pnas.2404887121. Epub 2024 Jul 5. Proc Natl Acad Sci U S A. 2024. PMID: 38968100 Free PMC article.
-
Prospects for the accelerated improvement of the resilient crop quinoa.J Exp Bot. 2020 Sep 19;71(18):5333-5347. doi: 10.1093/jxb/eraa285. J Exp Bot. 2020. PMID: 32643753 Free PMC article.
-
The MKK2a Gene Involved in the MAPK Signaling Cascades Enhances Populus Salt Tolerance.Int J Mol Sci. 2022 Sep 5;23(17):10185. doi: 10.3390/ijms231710185. Int J Mol Sci. 2022. PMID: 36077589 Free PMC article.
-
Genome-Wide Identification of MKK Gene Family and Response to Hormone and Abiotic Stress in Rice.Plants (Basel). 2024 Oct 18;13(20):2922. doi: 10.3390/plants13202922. Plants (Basel). 2024. PMID: 39458871 Free PMC article.
References
-
- Chono M, Matsunaka H, Seki M, Fujita M, Kiribuchi-Otobe C, Oda S, Kojima H, Nakamura S. Molecular and genealogical analysis of grain dormancy in Japanese wheat varieties, with specific focus on MOTHER OF FT AND TFL1 on chromosome 3A. Breed Sci. 2015;65:103–109. doi: 10.1270/jsbbs.65.103. - DOI - PMC - PubMed
-
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413:217–226 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

