Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus
- PMID: 30671943
- PMCID: PMC6554043
- DOI: 10.1002/ijc.32152
Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus
Abstract
Over the past decade, there has been a change in the epidemiology of oral cavity squamous cell cancer (OC-SCC). Many new cases of OC-SCC lack the recognized risk factors of smoking, alcohol and human papilloma virus. The aim of this study was to determine if the oral microbiome may be associated with OC-SCC in nonsmoking HPV negative patients. We compared the oral microbiome of HPV-negative nonsmoker OC-SCC(n = 18), premalignant lesions(PML) (n = 8) and normal control patients (n = 12). Their oral microbiome was sampled by oral wash and defined by 16S rRNA gene sequencing. We report that the periodontal pathogens Fusobacterium, Prevotella, Alloprevotella were enriched while commensal Streptococcus depleted in OC-SCC. Based on the four genera plus a marker genus Veillonella for PML, we classified the oral microbiome into two types. Gene/pathway analysis revealed a progressive increase of genes encoding HSP90 and ligands for TLRs 1, 2 and 4 along the controls→PML → OC-SCC progression sequence. Our findings suggest an association between periodontal pathogens and OC-SCC in non smoking HPV negative patients.
Keywords: Fusobacterium; streptococcus; leukoplakia; microbiome; nonsmoking; oral cavity squamous cell carcinoma; periodontal pathogens; risk factor.
© 2019 UICC.
Conflict of interest statement
Figures


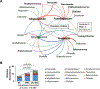
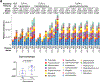
Similar articles
-
Association of Oral Microbiome With Risk for Incident Head and Neck Squamous Cell Cancer.JAMA Oncol. 2018 Mar 1;4(3):358-365. doi: 10.1001/jamaoncol.2017.4777. JAMA Oncol. 2018. PMID: 29327043 Free PMC article.
-
Epidemiology and Patient Distribution of Oral Cavity and Oropharyngeal SCC in Canada.J Cutan Med Surg. 2020 Jul/Aug;24(4):340-349. doi: 10.1177/1203475420915448. Epub 2020 Apr 2. J Cutan Med Surg. 2020. PMID: 32238063
-
Oral Microbiome in Nonsmoker Patients with Oral Cavity Squamous Cell Carcinoma, Defined by Metagenomic Shotgun Sequencing.Cancers (Basel). 2022 Dec 11;14(24):6096. doi: 10.3390/cancers14246096. Cancers (Basel). 2022. PMID: 36551584 Free PMC article.
-
The prevalence of human papilloma virus (HPV) infections in oral squamous cell carcinomas: a retrospective analysis of 88 patients and literature overview.J Craniomaxillofac Surg. 2014 Oct;42(7):1506-14. doi: 10.1016/j.jcms.2014.04.022. Epub 2014 May 10. J Craniomaxillofac Surg. 2014. PMID: 24947612 Review.
-
An Integrated Approach for Preventing Oral Cavity and Oropharyngeal Cancers: Two Etiologies with Distinct and Shared Mechanisms of Carcinogenesis.Cancer Prev Res (Phila). 2020 Aug;13(8):649-660. doi: 10.1158/1940-6207.CAPR-20-0096. Epub 2020 May 20. Cancer Prev Res (Phila). 2020. PMID: 32434808 Free PMC article. Review.
Cited by
-
Mechanisms and Potential Clinical Implications of Oral Microbiome in Oral Squamous Cell Carcinoma.Curr Oncol. 2023 Dec 28;31(1):168-182. doi: 10.3390/curroncol31010011. Curr Oncol. 2023. PMID: 38248096 Free PMC article. Review.
-
Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer.Technol Cancer Res Treat. 2019 Jan 1;18:1533033819867354. doi: 10.1177/1533033819867354. Technol Cancer Res Treat. 2019. PMID: 31370775 Free PMC article. Review.
-
Oral Microbiome Community Composition in Head and Neck Squamous Cell Carcinoma.Cancers (Basel). 2023 Apr 29;15(9):2549. doi: 10.3390/cancers15092549. Cancers (Basel). 2023. PMID: 37174014 Free PMC article.
-
Porphyromonas gingivalis upregulates calbindin 1 and thus promotes the proliferation of gingival epithelial cells.Hua Xi Kou Qiang Yi Xue Za Zhi. 2022 Jan 25;40(1):93-99. doi: 10.7518/hxkq.2022.01.014. Hua Xi Kou Qiang Yi Xue Za Zhi. 2022. PMID: 38596999 Free PMC article. Chinese, English.
-
Exploring the role of oral bacteria in oral cancer: a narrative review.Discov Oncol. 2025 Feb 26;16(1):242. doi: 10.1007/s12672-025-01998-2. Discov Oncol. 2025. PMID: 40009328 Free PMC article. Review.
References
-
- Wen BW, Tsai CS, Lin CL, et al. Cancer risk among gingivitis and periodontitis patients: a nationwide cohort study. Qjm-an International Journal of Medicine 2014;107: 283–290. - PubMed
-
- Tezal M, Sullivan MA, Reid ME, et al. Chronic periodontitis and the risk of tongue cancer. Archives of Otolaryngology - Head and Neck Surgery 2007;133: 450–454. - PubMed
-
- Zheng TZ, Boyle P, Hu HF, et al. Dentition, oral hygiene, and risk of oral cancer: a case-control study in Beijing, People’s Republic of China. Cancer Causes and Control 1990;1: 235–241. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

