Genome constellations of 24 porcine rotavirus group A strains circulating on commercial Thai swine farms between 2011 and 2016
- PMID: 30673764
- PMCID: PMC6343967
- DOI: 10.1371/journal.pone.0211002
Genome constellations of 24 porcine rotavirus group A strains circulating on commercial Thai swine farms between 2011 and 2016
Abstract
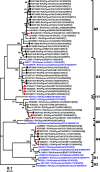
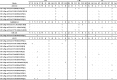
Rotavirus A (RVA) infection is a major cause of diarrhea-related illness in young children. RVA is also one of the most common enteric viruses detected on pig farms and contributes to substantial morbidity and mortality in piglets. Long-term multi-site surveillance of RVA on Thai swine farms to determine the diversity of RVA strains in circulation is currently lacking. In this study, we characterized the 11 segments of the RVA genome from 24 Thai porcine RVA strains circulating between 2011 and 2016. We identified G9 (15/24) and P[13] (12/24) as the dominant genotypes. The dominant G and P combinations were G9P[13] (n = 6), G9P[23] (n = 6), G3P[13] (n = 5), G9P[19] (n = 3), G4P[6] (n = 2), G4P[19] (n = 1), and G5P[13] (n = 1). Genome constellation of the Thai strains showed the predominance of Wa-like genotype (Gx-P[x]-I1/I5-R1-C1-M1-A8-N1-T1/T7-E1/E9-H1) with evidence of reassortment between the porcine and human RVA strains (e.g., G4-P[6]-I1-R1-C1-M1-A8-N1-T1-E1-H1 and G9-P[19]-I5-R1-C1-M1-A8-N1-T7-E9-H1). To assess the potential effectiveness of rotavirus vaccination, the Thai RVA strains were compared to the RVA strains represented in the swine rotavirus vaccine, which showed residue variations in the antigenic epitope on VP7 and shared amino acid identity below 90% for G4 and G5 strain. Several previous studies suggested these variations might effect on virus neutralization specificity and vaccine efficacy. Our study illustrates the importance of RVA surveillance beyond the G/P genotyping on commercial swine farms, which is crucial for controlling viral transmission.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Detection and genetic characterization of porcine group A rotaviruses in asymptomatic pigs in smallholder farms in East Africa: predominance of P[8] genotype resembling human strains.Vet Microbiol. 2015 Feb 25;175(2-4):195-210. doi: 10.1016/j.vetmic.2014.11.027. Epub 2014 Dec 10. Vet Microbiol. 2015. PMID: 25541378
-
Metagenomic sequencing generates the whole genomes of porcine rotavirus A, C, and H from the United States.PLoS One. 2020 Dec 29;15(12):e0244498. doi: 10.1371/journal.pone.0244498. eCollection 2020. PLoS One. 2020. PMID: 33373390 Free PMC article.
-
H2 genotypes of G4P[6], G5P[7], and G9[23] porcine rotaviruses show super-short RNA electropherotypes.Vet Microbiol. 2015 Apr 17;176(3-4):250-6. doi: 10.1016/j.vetmic.2015.02.002. Epub 2015 Feb 9. Vet Microbiol. 2015. PMID: 25724331
-
Porcine group A rotaviruses with heterogeneous VP7 and VP4 genotype combinations can be found together with enteric bacteria on Belgian swine farms.Vet Microbiol. 2014 Aug 6;172(1-2):23-34. doi: 10.1016/j.vetmic.2014.04.002. Epub 2014 Apr 13. Vet Microbiol. 2014. PMID: 24837191 Review.
-
Genotype constellation and evolution of group A rotaviruses infecting humans.Curr Opin Virol. 2012 Aug;2(4):426-33. doi: 10.1016/j.coviro.2012.04.007. Epub 2012 Jun 9. Curr Opin Virol. 2012. PMID: 22683209 Review.
Cited by
-
Whole-genome analysis of rotavirus G4P[6] strains isolated from Korean neonates: association of Korean neonates and rotavirus P[6] genotypes.Gut Pathog. 2019 Jul 10;11:37. doi: 10.1186/s13099-019-0318-5. eCollection 2019. Gut Pathog. 2019. PMID: 31333764 Free PMC article.
-
Genetic Diversity of Porcine Group A Rotavirus Strains from Pigs in South Korea.Viruses. 2022 Nov 14;14(11):2522. doi: 10.3390/v14112522. Viruses. 2022. PMID: 36423131 Free PMC article.
-
Driving forces of continuing evolution of rotaviruses.World J Virol. 2024 Jun 25;13(2):93774. doi: 10.5501/wjv.v13.i2.93774. World J Virol. 2024. PMID: 38984077 Free PMC article. Review.
-
Genetic Diversity of Rotaviruses Circulating in Pediatric Patients and Domestic Animals in Thailand.Trop Med Infect Dis. 2023 Jun 29;8(7):347. doi: 10.3390/tropicalmed8070347. Trop Med Infect Dis. 2023. PMID: 37505643 Free PMC article. Review.
-
Rotavirus Infection in Swine: Genotypic Diversity, Immune Responses, and Role of Gut Microbiome in Rotavirus Immunity.Pathogens. 2022 Sep 22;11(10):1078. doi: 10.3390/pathogens11101078. Pathogens. 2022. PMID: 36297136 Free PMC article. Review.
References
-
- Pesavento JB, Crawford SE, Estes MK, Prasad BV. Rotavirus proteins: structure and assembly. Curr Top Microbiol Immunol. 2006;309:189–219. Epub 2006/08/18. . - PubMed
-
- Estes M.K and Greenberg HB. Rotaviruses In: Knipe DM, Howley P.M., et al. (Eds.), editor. Fields virology. sixth ed Philadelphia: Wolters Kluwer—Lippincott Williams & Wilkins; 2013. p. 1347–401.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

