Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus
- PMID: 30673820
- PMCID: PMC11105500
- DOI: 10.1007/s00018-019-03021-8
Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus
Abstract
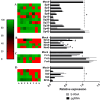
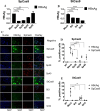
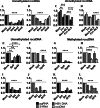
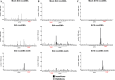
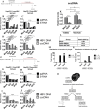
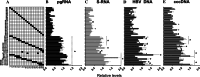
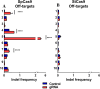
Covalently closed circular DNA (cccDNA) of hepatitis B virus (HBV) is the major cause of viral persistence and chronic hepatitis B. CRISPR/Cas9 nucleases can specifically target HBV cccDNA for decay, but off-target effects of nucleases in the human genome limit their clinical utility. CRISPR/Cas9 systems from four different species were co-expressed in cell lines with guide RNAs targeting conserved regions of the HBV genome. CRISPR/Cas9 systems from Streptococcus pyogenes (Sp) and Streptococcus thermophilus (St) targeting conserved regions of the HBV genome blocked HBV replication and, most importantly, resulted in degradation of over 90% of HBV cccDNA by 6 days post-transfection. Degradation of HBV cccDNA was impaired by inhibition of non-homologous end-joining pathway and resulted in an erroneous repair of HBV cccDNA. HBV cccDNA methylation also affected antiviral activity of CRISPR/Cas9. Single-nucleotide HBV genetic variants did not impact anti-HBV activity of St CRISPR/Cas9, suggesting its utility in targeting many HBV variants. However, two or more mismatches impaired or blocked CRISPR/Cas9 activity, indicating that host DNA will not likely be targeted. Deep sequencing revealed that Sp CRISPR/Cas9 induced off-target mutagenesis, whereas St CRISPR/Cas9 had no effect on the host genome. St CRISPR/Cas9 system represents the safest system with high anti-HBV activity.
Keywords: Antiviral; Cure; Liver; Mutations; NHEJ; Therapeutics.
Conflict of interest statement
The authors declare that they have no potential conflicts of interests. The authors have applied for patents concerning the use of Cas9 proteins and gRNAs for HBV therapy.
Figures
References
-
- World Health Organization . Global hepatitis report 2017. Geneva: World Health Organization; 2017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

