Sleeping Beauty Insertional Mutagenesis Reveals Important Genetic Drivers of Central Nervous System Embryonal Tumors
- PMID: 30674530
- PMCID: PMC6397665
- DOI: 10.1158/0008-5472.CAN-18-1261
Sleeping Beauty Insertional Mutagenesis Reveals Important Genetic Drivers of Central Nervous System Embryonal Tumors
Abstract
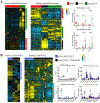
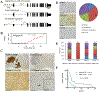
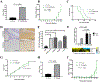
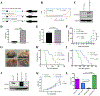
Medulloblastoma and central nervous system primitive neuroectodermal tumors (CNS-PNET) are aggressive, poorly differentiated brain tumors with limited effective therapies. Using Sleeping Beauty (SB) transposon mutagenesis, we identified novel genetic drivers of medulloblastoma and CNS-PNET. Cross-species gene expression analyses classified SB-driven tumors into distinct medulloblastoma and CNS-PNET subgroups, indicating they resemble human Sonic hedgehog and group 3 and 4 medulloblastoma and CNS neuroblastoma with FOXR2 activation. This represents the first genetically induced mouse model of CNS-PNET and a rare model of group 3 and 4 medulloblastoma. We identified several putative proto-oncogenes including Arhgap36, Megf10, and Foxr2. Genetic manipulation of these genes demonstrated a robust impact on tumorigenesis in vitro and in vivo. We also determined that FOXR2 interacts with N-MYC, increases C-MYC protein stability, and activates FAK/SRC signaling. Altogether, our study identified several promising therapeutic targets in medulloblastoma and CNS-PNET. SIGNIFICANCE: A transposon-induced mouse model identifies several novel genetic drivers and potential therapeutic targets in medulloblastoma and CNS-PNET.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures


References
-
- Chan TSY, Wang X, Spence T, Taylor MD, Huang A Embryonal brain tumors. New York: Springer; 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

