JACKS: joint analysis of CRISPR/Cas9 knockout screens
- PMID: 30674557
- PMCID: PMC6396427
- DOI: 10.1101/gr.238923.118
JACKS: joint analysis of CRISPR/Cas9 knockout screens
Abstract
Genome-wide CRISPR/Cas9 knockout screens are revolutionizing mammalian functional genomics. However, their range of applications remains limited by signal variability from different guide RNAs that target the same gene, which confounds gene effect estimation and dictates large experiment sizes. To address this problem, we report JACKS, a Bayesian method that jointly analyzes screens performed with the same guide RNA library. Modeling the variable guide efficacies greatly improves hit identification over processing a single screen at a time and outperforms existing methods. This more efficient analysis gives additional hits and allows designing libraries with a 2.5-fold reduction in required cell numbers without sacrificing performance compared to current analysis standards.
© 2019 Allen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
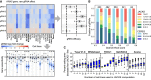
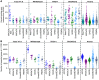


References
-
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. 2016. Genomic copy number dictates a gene-independent cell response to CRISPR-Cas9 targeting. Cancer Discov 6: 914–929. 10.1158/2159-8290.CD-16-0154 - DOI - PMC - PubMed
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300. 10.2307/2346101 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
