Potent Anti-hepatitis C Virus (HCV) T Cell Immune Responses Induced in Mice Vaccinated with DNA-Launched RNA Replicons and Modified Vaccinia Virus Ankara-HCV
- PMID: 30674625
- PMCID: PMC6430543
- DOI: 10.1128/JVI.00055-19
Potent Anti-hepatitis C Virus (HCV) T Cell Immune Responses Induced in Mice Vaccinated with DNA-Launched RNA Replicons and Modified Vaccinia Virus Ankara-HCV
Abstract
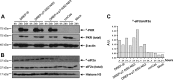
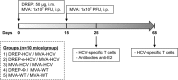
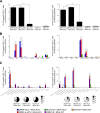
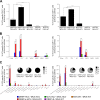
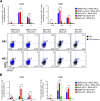
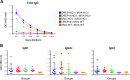
Hepatitis C is a liver disease caused by the hepatitis C virus (HCV) affecting 71 million people worldwide with no licensed vaccines that prevent infection. Here, we have generated four novel alphavirus-based DNA-launched self-amplifying RNA replicon (DREP) vaccines expressing either structural core-E1-E2 or nonstructural p7-NS2-NS3 HCV proteins of genotype 1a placed under the control of an alphavirus promoter, with or without an alphaviral translational enhancer (grouped as DREP-HCV or DREP-e-HCV, respectively). DREP vectors are known to induce cross-priming and further stimulation of immune responses through apoptosis, and here we demonstrate that they efficiently trigger apoptosis-related proteins in transfected cells. Immunization of mice with the DREP vaccines as the priming immunization followed by a heterologous boost with a recombinant modified vaccinia virus Ankara (MVA) vector expressing the nearly full-length genome of HCV (MVA-HCV) induced potent and long-lasting HCV-specific CD4+ and CD8+ T cell immune responses that were significantly stronger than those of a homologous MVA-HCV prime/boost immunization, with the DREP-e-HCV/MVA-HCV combination the most immunogenic regimen. HCV-specific CD4+ and CD8+ T cell responses were highly polyfunctional, had an effector memory phenotype, and were mainly directed against E1-E2 and NS2-NS3, respectively. Additionally, DREP/MVA-HCV immunization regimens induced higher antibody levels against HCV E2 protein than homologous MVA-HCV immunization. Collectively, these results provided an immunization protocol against HCV by inducing high levels of HCV-specific T cell responses as well as humoral responses. These findings reinforce the combined use of DREP-based vectors and MVA-HCV as promising prophylactic and therapeutic vaccines against HCV.IMPORTANCE HCV represents a global health problem as more than 71 million people are chronically infected worldwide. Direct-acting antiviral agents can cure HCV infection in most patients, but due to the high cost of these agents and the emergence of resistant mutants, they do not represent a feasible and affordable strategy to eradicate the virus. Therefore, a vaccine is an urgent goal that requires efforts to understand the correlates of protection for HCV clearance. Here, we describe for the first time the generation of novel vaccines against HCV based on alphavirus DNA replicons expressing HCV antigens. We demonstrate that potent T cell immune responses, as well as humoral immune responses, against HCV can be achieved in mice by using a combined heterologous prime/boost immunization protocol consisting of the administration of alphavirus replicon DNA vectors as the priming immunization followed by a boost with a recombinant modified vaccinia virus Ankara vector expressing HCV antigens.
Keywords: HCV; MVA; alphavirus replicon; immune response; mice; poxvirus; vaccine.
Copyright © 2019 American Society for Microbiology.
Figures

Similar articles
-
Alphavirus replicon DNA expressing HIV antigens is an excellent prime for boosting with recombinant modified vaccinia Ankara (MVA) or with HIV gp140 protein antigen.PLoS One. 2015 Feb 2;10(2):e0117042. doi: 10.1371/journal.pone.0117042. eCollection 2015. PLoS One. 2015. PMID: 25643354 Free PMC article.
-
DNA-launched RNA replicon vaccines induce potent anti-Ebolavirus immune responses that can be further improved by a recombinant MVA boost.Sci Rep. 2018 Aug 20;8(1):12459. doi: 10.1038/s41598-018-31003-6. Sci Rep. 2018. PMID: 30127450 Free PMC article.
-
High, broad, polyfunctional, and durable T cell immune responses induced in mice by a novel hepatitis C virus (HCV) vaccine candidate (MVA-HCV) based on modified vaccinia virus Ankara expressing the nearly full-length HCV genome.J Virol. 2013 Jul;87(13):7282-300. doi: 10.1128/JVI.03246-12. Epub 2013 Apr 17. J Virol. 2013. PMID: 23596307 Free PMC article.
-
Recombinant MVA vaccines: dispelling the myths.Vaccine. 2013 Sep 6;31(39):4247-51. doi: 10.1016/j.vaccine.2013.03.021. Epub 2013 Mar 21. Vaccine. 2013. PMID: 23523407 Review.
-
Modified Vaccinia Virus Ankara: History, Value in Basic Research, and Current Perspectives for Vaccine Development.Adv Virus Res. 2017;97:187-243. doi: 10.1016/bs.aivir.2016.07.001. Epub 2016 Aug 1. Adv Virus Res. 2017. PMID: 28057259 Free PMC article. Review.
Cited by
-
Application of DNA Replicons in Gene Therapy and Vaccine Development.Pharmaceutics. 2023 Mar 15;15(3):947. doi: 10.3390/pharmaceutics15030947. Pharmaceutics. 2023. PMID: 36986808 Free PMC article. Review.
-
COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike induce robust T- and B-cell immune responses and full efficacy in mice.J Virol. 2021 Mar 10;95(7):e02260-20. doi: 10.1128/JVI.02260-20. Epub 2021 Jan 7. J Virol. 2021. PMID: 33414159 Free PMC article.
-
Optimized Hepatitis C Virus (HCV) E2 Glycoproteins and their Immunogenicity in Combination with MVA-HCV.Vaccines (Basel). 2020 Aug 5;8(3):440. doi: 10.3390/vaccines8030440. Vaccines (Basel). 2020. PMID: 32764419 Free PMC article.
-
Contemporary Insights into Hepatitis C Virus: A Comprehensive Review.Microorganisms. 2024 May 21;12(6):1035. doi: 10.3390/microorganisms12061035. Microorganisms. 2024. PMID: 38930417 Free PMC article. Review.
-
The combined vaccination protocol of DNA/MVA expressing Zika virus structural proteins as efficient inducer of T and B cell immune responses.Emerg Microbes Infect. 2021 Dec;10(1):1441-1456. doi: 10.1080/22221751.2021.1951624. Emerg Microbes Infect. 2021. PMID: 34213405 Free PMC article.
References
-
- Bartenschlager R, Baumert TF, Bukh J, Houghton M, Lemon SM, Lindenbach BD, Lohmann V, Moradpour D, Pietschmann T, Rice CM, Thimme R, Wakita T. 2018. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res 248:53–62. doi:10.1016/j.virusres.2018.02.016. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

