An improved yeast surface display platform for the screening of nanobody immune libraries
- PMID: 30674983
- PMCID: PMC6344588
- DOI: 10.1038/s41598-018-37212-3
An improved yeast surface display platform for the screening of nanobody immune libraries
Abstract
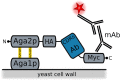
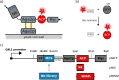
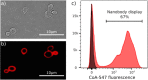
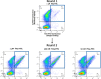
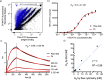
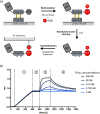
Fusions to the C-terminal end of the Aga2p mating adhesion of Saccharomyces cerevisiae have been used in many studies for the selection of affinity reagents by yeast display followed by flow cytometric analysis. Here we present an improved yeast display system for the screening of Nanobody immune libraries where we fused the Nanobody to the N-terminal end of Aga2p to avoid steric hindrance between the fused Nanobody and the antigen. Moreover, the display level of a cloned Nanobody on the surface of an individual yeast cell can be monitored through a covalent fluorophore that is attached in a single enzymatic step to an orthogonal acyl carrier protein (ACP). Additionally, the displayed Nanobody can be easily released from the yeast surface and immobilised on solid surfaces for rapid analysis. To prove the generic nature of this novel Nanobody discovery platform, we conveniently selected Nanobodies against three different antigens, including two membrane proteins.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

