Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid
- PMID: 30675060
- PMCID: PMC6457907
- DOI: 10.1038/s41586-019-0882-3
Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid
Abstract
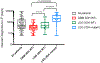
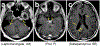
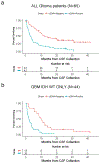
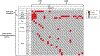
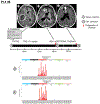
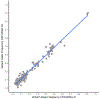
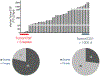
Diffuse gliomas are the most common malignant brain tumours in adults and include glioblastomas and World Health Organization (WHO) grade II and grade III tumours (sometimes referred to as lower-grade gliomas). Genetic tumour profiling is used to classify disease and guide therapy1,2, but involves brain surgery for tissue collection; repeated tumour biopsies may be necessary for accurate genotyping over the course of the disease3-10. While the detection of circulating tumour DNA (ctDNA) in the blood of patients with primary brain tumours remains challenging11,12, sequencing of ctDNA from the cerebrospinal fluid (CSF) may provide an alternative way to genotype gliomas with lower morbidity and cost13,14. We therefore evaluated the representation of the glioma genome in CSF from 85 patients with gliomas who underwent a lumbar puncture because they showed neurological signs or symptoms. Here we show that tumour-derived DNA was detected in CSF from 42 out of 85 patients (49.4%) and was associated with disease burden and adverse outcome. The genomic landscape of glioma in the CSF included a broad spectrum of genetic alterations and closely resembled the genomes of tumour biopsies. Alterations that occur early during tumorigenesis, such as co-deletion of chromosome arms 1p and 19q (1p/19q codeletion) and mutations in the metabolic genes isocitrate dehydrogenase 1 (IDH1) or IDH21,2, were shared in all matched ctDNA-positive CSF-tumour pairs, whereas growth factor receptor signalling pathways showed considerable evolution. The ability to monitor the evolution of the glioma genome through a minimally invasive technique could advance the clinical development and use of genotype-directed therapies for glioma, one of the most aggressive human cancers.
Figures



Comment in
-
Characterizing Glioma Genetics Using Cerebrospinal Fluid.Neurosurgery. 2019 Aug 1;85(2):E196-E197. doi: 10.1093/neuros/nyz177. Neurosurgery. 2019. PMID: 31304547 No abstract available.
-
Predicting Outcome using Genomic-Based Liquid Biomarkers.Int J Radiat Oncol Biol Phys. 2020 Jan 1;106(1):1-4. doi: 10.1016/j.ijrobp.2019.10.017. Int J Radiat Oncol Biol Phys. 2020. PMID: 31836074 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

