Features of a novel protein, rusticalin, from the ascidian Styela rustica reveal ancestral horizontal gene transfer event
- PMID: 30675192
- PMCID: PMC6339383
- DOI: 10.1186/s13100-019-0146-7
Features of a novel protein, rusticalin, from the ascidian Styela rustica reveal ancestral horizontal gene transfer event
Abstract
Background: The transfer of genetic material from non-parent organisms is called horizontal gene transfer (HGT). One of the most conclusive cases of HGT in metazoans was previously described for the cellulose synthase gene in ascidians.
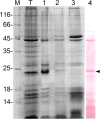
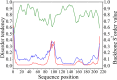
Results: In this study we identified a new protein, rusticalin, from the ascidian Styela rustica and presented evidence for its likely origin by HGT. Discernible homologues of rusticalin were found in placozoans, coral, and basal Chordates. Rusticalin was predicted to consist of two distinct regions, an N-terminal domain and a C-terminal domain. The N-terminal domain comprises two cysteine-rich repeats and shows remote similarity to the tick carboxypeptidase inhibitor. The C-terminal domain shares significant sequence similarity with bacterial MD peptidases and bacteriophage A500 L-alanyl-D-glutamate peptidase. A possible transfer of the C-terminal domain by bacteriophage was confirmed by an analysis of noncoding sequences of C. intestinalis rusticalin-like gene, which was found to contain a sequence similar to the bacteriophage A500 recombination site. Moreover, a sequence similar to the bacteriophage recombination site was found to be adjacent to the cellulose synthase catalytic subunit gene in the genome of Streptomices sp., the donor of ascidian cellulose synthase.
Conclusions: The C-terminal domain of rusticalin and rusticalin-like proteins is likely to be horizontally transferred by the bacteriophage A500. A common mechanism involving bacteriophage mediated gene transfer can be proposed for at least two HGT events in ascidians.
Keywords: Ascidians; Bacteriophage; Hemocytes; Horizontal gene transfer; L-alanyl-D-glutamate peptidase; Trichoplax; tRNA.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures










Similar articles
-
Amino acid sequence associated with bacteriophage recombination site helps to reveal genes potentially acquired through horizontal gene transfer.BMC Bioinformatics. 2020 Jul 24;21(Suppl 12):305. doi: 10.1186/s12859-020-03599-y. BMC Bioinformatics. 2020. PMID: 32703190 Free PMC article.
-
Transposon-Mediated Horizontal Transfer of the Host-Specific Virulence Protein ToxA between Three Fungal Wheat Pathogens.mBio. 2019 Sep 10;10(5):e01515-19. doi: 10.1128/mBio.01515-19. mBio. 2019. PMID: 31506307 Free PMC article.
-
A functional cellulose synthase from ascidian epidermis.Proc Natl Acad Sci U S A. 2004 Jan 27;101(4):986-91. doi: 10.1073/pnas.0303623101. Epub 2004 Jan 13. Proc Natl Acad Sci U S A. 2004. PMID: 14722352 Free PMC article.
-
Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches.FEMS Microbiol Rev. 2011 Sep;35(5):957-76. doi: 10.1111/j.1574-6976.2011.00292.x. Epub 2011 Jul 29. FEMS Microbiol Rev. 2011. PMID: 21711367 Review.
-
Horizontal Gene Transfer Contributes to Plant Evolution: The Case of Agrobacterium T-DNAs.Front Plant Sci. 2017 Nov 24;8:2015. doi: 10.3389/fpls.2017.02015. eCollection 2017. Front Plant Sci. 2017. PMID: 29225610 Free PMC article. Review.
Cited by
-
Amino acid sequence associated with bacteriophage recombination site helps to reveal genes potentially acquired through horizontal gene transfer.BMC Bioinformatics. 2020 Jul 24;21(Suppl 12):305. doi: 10.1186/s12859-020-03599-y. BMC Bioinformatics. 2020. PMID: 32703190 Free PMC article.
References
-
- Daele Y, Revol J, Gaill F, Goffinet G. Characterization and supramolecular architecture of the cellulose-protein fibrils in the tunic of the sea peach (Halocynthia papillosa, Ascidiacea, Urochordata) Biol Cell. 1992;76:87–96. doi: 10.1016/0248-4900(92)90198-A. - DOI
LinkOut - more resources
Full Text Sources

