Both rare and common genetic variants contribute to autism in the Faroe Islands
- PMID: 30675382
- PMCID: PMC6341098
- DOI: 10.1038/s41525-018-0075-2
Both rare and common genetic variants contribute to autism in the Faroe Islands
Abstract
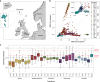
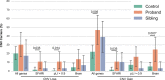
The number of genes associated with autism is increasing, but few studies have been performed on epidemiological cohorts and in isolated populations. Here, we investigated 357 individuals from the Faroe Islands including 36 individuals with autism, 136 of their relatives and 185 non-autism controls. Data from SNP array and whole exome sequencing revealed that individuals with autism had a higher burden of rare exonic copy-number variants altering autism associated genes (deletions (p = 0.0352) or duplications (p = 0.0352)), higher inbreeding status (p = 0.023) and a higher load of rare homozygous deleterious variants (p = 0.011) compared to controls. Our analysis supports the role of several genes/loci associated with autism (e.g., NRXN1, ADNP, 22q11 deletion) and identified new truncating (e.g., GRIK2, ROBO1, NINL, and IMMP2L) or recessive deleterious variants (e.g., KIRREL3 and CNTNAP2) affecting autism-associated genes. It also revealed three genes involved in synaptic plasticity, RIMS4, KALRN, and PLA2G4A, carrying de novo deleterious variants in individuals with autism without intellectual disability. In summary, our analysis provides a better understanding of the genetic architecture of autism in isolated populations by highlighting the role of both common and rare gene variants and pointing at new autism-risk genes. It also indicates that more knowledge about how multiple genetic hits affect neuronal function will be necessary to fully understand the genetic architecture of autism.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism. 2017;8:21. doi: 10.1186/s13229-017-0137-9. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

