Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury
- PMID: 30676324
- PMCID: PMC6478413
- DOI: 10.1172/jci.insight.124522
Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury
Abstract
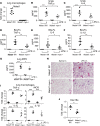
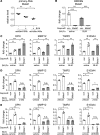
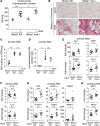
Macrophage activation, i.e., classical M1 and the alternative M2, plays a critical role in many pathophysiological processes, such as inflammation and tissue injury and repair. Although the regulation of macrophage activation has been under extensive investigation, there is little knowledge about the role of long noncoding RNAs (lncRNAs) in this event. In this study, we found that lncRNA Malat1 expression is distinctly regulated in differentially activated macrophages in that it is upregulated in LPS-treated and downregulated in IL-4-treated cells. Malat1 knockdown attenuates LPS-induced M1 macrophage activation. In contrast, Malat1 knockdown enhanced IL-4-activated M2 differentiation as well as a macrophage profibrotic phenotype. Mechanistically, Malat1 knockdown led to decreased expression of Clec16a, silencing of which phenocopied the regulatory effect of Malat1 on M1 activation. Interestingly, Malat1 knockdown promoted IL-4 induction of mitochondrial pyruvate carriers (MPCs) and their mediation of glucose-derived oxidative phosphorylation (OxPhos), which was crucial to the Malat1 regulation of M2 differentiation and profibrotic phenotype. Furthermore, mice with either global or conditional myeloid knockout of Malat1 demonstrated diminished LPS-induced systemic and pulmonary inflammation and injury. In contrast, these mice developed more severe bleomycin-induced lung fibrosis, accompanied by alveolar macrophages displaying augmented M2 and profibrotic phenotypes. In summary, we have identified what we believe is a previously unrecognized role of Malat1 in the regulation of macrophage polarization. Our data demonstrate that Malat1 is involved in pulmonary pathogeneses in association with aberrant macrophage activation.
Keywords: Cellular immune response; Fibrosis; Immunology; Macrophages; Pulmonology.
Conflict of interest statement
Figures





Similar articles
-
Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up-regulating miR-146a in LPS-induced acute lung injury.Connect Tissue Res. 2018 Nov;59(6):581-592. doi: 10.1080/03008207.2018.1439480. Epub 2018 Apr 13. Connect Tissue Res. 2018. PMID: 29649906
-
Influence of lncRNA MALAT1 on septic lung injury in mice through p38 MAPK/p65 NF-κB pathway.Eur Rev Med Pharmacol Sci. 2019 Feb;23(3):1296-1304. doi: 10.26355/eurrev_201902_17025. Eur Rev Med Pharmacol Sci. 2019. PMID: 30779099
-
Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice.J Immunol. 2014 Jan 1;192(1):394-406. doi: 10.4049/jimmunol.1300959. Epub 2013 Nov 25. J Immunol. 2014. PMID: 24277697
-
The roles of long noncoding RNA-mediated macrophage polarization in respiratory diseases.Front Immunol. 2023 Jan 5;13:1110774. doi: 10.3389/fimmu.2022.1110774. eCollection 2022. Front Immunol. 2023. PMID: 36685535 Free PMC article. Review.
-
LncRNA MALAT1 as a potential diagnostic and therapeutic target in kidney diseases.Pathol Res Pract. 2025 Feb;266:155783. doi: 10.1016/j.prp.2024.155783. Epub 2024 Dec 20. Pathol Res Pract. 2025. PMID: 39724850 Review.
Cited by
-
Role and mechanism of ferroptosis in acute lung injury.Cell Cycle. 2023 Oct;22(19):2119-2129. doi: 10.1080/15384101.2023.2278328. Epub 2023 Dec 5. Cell Cycle. 2023. PMID: 37946318 Free PMC article. Review.
-
Malat1 regulates PMN-MDSC expansion and immunosuppression through p-STAT3 ubiquitination in sepsis.Int J Biol Sci. 2024 Feb 11;20(4):1529-1546. doi: 10.7150/ijbs.92267. eCollection 2024. Int J Biol Sci. 2024. PMID: 38385073 Free PMC article.
-
LncRNAs: The Regulator of Glucose and Lipid Metabolism in Tumor Cells.Front Oncol. 2019 Nov 26;9:1099. doi: 10.3389/fonc.2019.01099. eCollection 2019. Front Oncol. 2019. PMID: 31850189 Free PMC article. Review.
-
Long non-coding RNAs (lncRNAs) NEAT1 and MALAT1 are differentially expressed in severe COVID-19 patients: An integrated single-cell analysis.PLoS One. 2022 Jan 10;17(1):e0261242. doi: 10.1371/journal.pone.0261242. eCollection 2022. PLoS One. 2022. PMID: 35007307 Free PMC article.
-
Suppression of the JAK/STAT Pathway Inhibits Neuroinflammation in the Line 61-PFF Mouse Model of Parkinson's Disease.Res Sq [Preprint]. 2024 May 7:rs.3.rs-4307273. doi: 10.21203/rs.3.rs-4307273/v1. Res Sq. 2024. Update in: J Neuroinflammation. 2024 Sep 1;21(1):216. doi: 10.1186/s12974-024-03210-8. PMID: 38766241 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

