O-GlcNAc transferase activates stem-like cell potential in hepatocarcinoma through O-GlcNAcylation of eukaryotic initiation factor 4E
- PMID: 30677218
- PMCID: PMC6433694
- DOI: 10.1111/jcmm.14043
O-GlcNAc transferase activates stem-like cell potential in hepatocarcinoma through O-GlcNAcylation of eukaryotic initiation factor 4E
Abstract
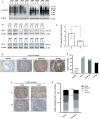
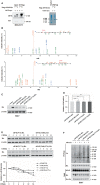
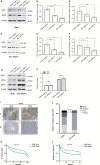
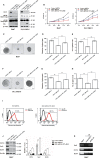
O-GlcNAcylation catalysed by O-GlcNAc transferase (OGT) is a reversible post-translational modification. O-GlcNAcylation participates in transcription, epigenetic regulation, and intracellular signalling. Dysregulation of O-GlcNAcylation in response to high glucose or OGT expression has been implicated in metabolic diseases and cancer. However, the underlying mechanisms by which OGT regulates hepatoma development remain largely unknown. Here, we employed the lentiviral shRNA-based system to knockdown OGT to analyse the contribution of OGT in hepatoma cell proliferation and stem-like cell potential. The sphere-forming assay and western blot analysis of stem-related gene expression were used to evaluate stem-like cell potential of hepatoma cell. We found that the level of total O-GlcNAcylation or OGT protein was increased in hepatocellular carcinoma. OGT activated stem-like cell potential in hepatoma through eukaryotic initiation factor 4E (eIF4E) which bound to stem-related gene Sox2 5'-untranslated region. O-GlcNAcylation of eIF4E at threonine 168 and threonine 177 protected it from degradation through proteasome pathway. Expression of eIF4E in hepatoma was determined by immunostaining in 232 HCC patients, and Kaplan-Meier survival analysis was used to determine the correlation of eIF4E expression with prognosis. High glucose promoted stem-like cell potential of hepatoma cell through OGT-eIF4E axis. Collectively, our findings indicate that OGT promotes the stem-like cell potential of hepatoma cell through O-GlcNAcylation of eIF4E. These results provide a mechanism of HCC development and a cue between the pathogenesis of HCC and high glucose condition.
Keywords: O-GlcNAc transferase; O-GlcNAcylation; eukaryotic initiation factor 4E; hepatocellular carcinoma; stem-like cell potential.
© 2018 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Wild CP, Hall AJ. Primary prevention of hepatocellular carcinoma in developing countries. Mutat Res. 2000;462:381‐393. - PubMed
-
- Fasani P, Sangiovanni A, De Fazio C, et al. High prevalence of multinodular hepatocellular carcinoma in patients with cirrhosis attributable to multiple risk factors. Hepatology. 1999;29:1704‐1707. - PubMed
-
- Lai S‐W, Chen P‐C, Liao K‐F, et al. Risk of hepatocellular carcinoma in diabetic patients and risk reduction associated with anti‐diabetic therapy: a population‐based cohort study. Am J Gastroenterol. 2011;107:46‐52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

