Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011-2017
- PMID: 30677534
- PMCID: PMC7106134
- DOI: 10.1016/j.meegid.2019.01.022
Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011-2017
Abstract
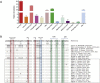
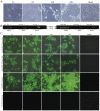
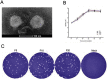
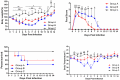
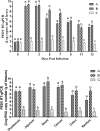
In recent years, the outbreaks of porcine epidemic diarrhea (PED) caused by the highly virulent porcine epidemic diarrhea virus (PEDV) variants occurred frequently in China, resulting in severe economic impacts to the pork industry. In this study, we selected and analyzed the genetic evolution of 15 PEDV representative strains that were identified in fecal samples of diarrheic piglets in 10 provinces and cities during 2011-2017. The phylogenetic analysis indicated that all the 15 PEDV isolates clustered into G2 genotype associated with the current circulating strains. Compared with the genome of the prototype strain CV777, these strains had 103-120 amino acid mutations in their S proteins, most of which were in the N terminal domain of S1 (S1-NTD). We also found 37 common mutations in all these 15 strains, although these strains shared 96.9-99.7% nucleotide homology and 96.3-99.8% amino acid homology in the S protein compared with the other original pandemic strains. Computational analysis showed that these mutations may lead to remarkable changes in the conformational structure and asparagine (N)-linked glycosylation sites of S1-NTD, which may be associated with the altered pathogenicity of these variant PEDV strains. We evaluated the pathogenicity of the PEDV strain FJzz1 in piglets through oral and intramuscular infection routes. Compared with oral infection, intramuscular infection could also cause typical clinical signs but with a slightly delayed onset, confirming that the variant PEDV isolate FJzz1 was highly pathogenic to suckling piglets. In conclusion, we analyzed the genetic variation and pathogenicity of the emerging PEDV isolates of China, indicating that G2 variant PEDV strains as the main prevalent strains that may mutate continually. This study shows the necessity of monitoring the molecular epidemiology and the etiological characteristics of the epidemic PEDV isolates, which may help better control the PED outbreaks.
Keywords: FJzz1; PEDV; Pathogenicity; S gene; Variation.
Copyright © 2019 Elsevier B.V. All rights reserved.
Figures



References
-
- Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

