Using Fluorescence Quenching Titration to Determine the Orientation of a Model Transmembrane Protein in Mimic Membranes
- PMID: 30678051
- PMCID: PMC6384929
- DOI: 10.3390/ma12030349
Using Fluorescence Quenching Titration to Determine the Orientation of a Model Transmembrane Protein in Mimic Membranes
Abstract
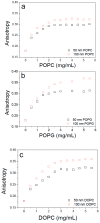
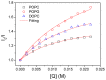
After synthesis of transmembrane proteins (TMPs), they are transferred and inserted into plasma membranes to play biological functions. Crucially, orientation of TMPs in membranes determines whether they have biological activities. In cellular environments, a number of cofactors, such as translocon, can assist TMPs to be inserted into membranes in defined orientations. During in vitro reconstitution of TMPs with mimic membranes, both insertion and orientation of TMPs are primarily determined by interactions with the membrane. Yet the knowledge is limited, hindering the in vitro applications of TMPs. Here, we take Bacteriorhodopsin (bR) as a model TMP, using fluorescence quenching titration experiment to identify orientation of bR in mimic membranes, examining effects of a number of factors, including lipid composition, pH value, ionic strength and membrane curvature. The most effective determinant is the lipid type, which modulates insertion and orientation of bR in membranes by changing the membrane surface charge and the membrane fluidity. Both the pH value and the ionic strength play secondary roles by tuning the nature of the electrostatic interaction. The membrane curvature was found to have a minor effect on orientation of bR in membranes. By comparing orientations of bR in folded and unfolded states, no obvious change was observed, informing that nascent proteins could be inserted into membranes in defined orientations before folding into the native state inside the membrane.
Keywords: fluorescence quenching titration; insertion; mimic membrane; orientation; transmembrane protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Folding of β-Barrel Membrane Proteins into Lipid Membranes by Site-Directed Fluorescence Spectroscopy.Methods Mol Biol. 2019;2003:465-492. doi: 10.1007/978-1-4939-9512-7_20. Methods Mol Biol. 2019. PMID: 31218630
-
How transmembrane peptides insert and orientate in biomembranes: a combined experimental and simulation study.Phys Chem Chem Phys. 2016 Jun 29;18(26):17483-94. doi: 10.1039/c6cp01133k. Phys Chem Chem Phys. 2016. PMID: 27302083
-
Kinetics of Insertion and Folding of Outer Membrane Proteins by Gel Electrophoresis.Methods Mol Biol. 2019;2003:145-162. doi: 10.1007/978-1-4939-9512-7_7. Methods Mol Biol. 2019. PMID: 31218617
-
AMPs and OMPs: Is the folding and bilayer insertion of β-stranded outer membrane proteins governed by the same biophysical principles as for α-helical antimicrobial peptides?Biochim Biophys Acta. 2015 Sep;1848(9):1944-54. doi: 10.1016/j.bbamem.2015.02.019. Epub 2015 Feb 27. Biochim Biophys Acta. 2015. PMID: 25726906 Review.
-
Functional and Biomimetic DNA Nanostructures on Lipid Membranes.Langmuir. 2018 Dec 11;34(49):14721-14730. doi: 10.1021/acs.langmuir.8b01818. Epub 2018 Sep 10. Langmuir. 2018. PMID: 30044097 Review.
Cited by
-
Emerging Designs and Applications for Biomembrane Biosensors.Annu Rev Anal Chem (Palo Alto Calif). 2024 Jul;17(1):339-366. doi: 10.1146/annurev-anchem-061622-042618. Annu Rev Anal Chem (Palo Alto Calif). 2024. PMID: 39018354 Free PMC article. Review.
-
Rapid Estimation of Membrane Protein Orientation in Liposomes.Chembiochem. 2022 Jan 19;23(2):e202100543. doi: 10.1002/cbic.202100543. Epub 2021 Nov 24. Chembiochem. 2022. PMID: 34763366 Free PMC article.
References
LinkOut - more resources
Full Text Sources

