Toll-Like Receptor-Dependent Immunomodulatory Activity of Pycnogenol®
- PMID: 30678156
- PMCID: PMC6412808
- DOI: 10.3390/nu11020214
Toll-Like Receptor-Dependent Immunomodulatory Activity of Pycnogenol®
Abstract
Background: Pycnogenol® (PYC), an extract of French maritime pine bark, is widely used as a dietary supplement. PYC has been shown to exert anti-inflammatory actions via inhibiting the Toll-like receptor 4 (TLR4) pathway. However, the role of the other receptors from the TLR family in the immunomodulatory activity of PYC has not been described so far.
Aim: The aim of this study was to investigate whether PYC might exert its immunomodulatory properties through cell membrane TLRs (TLR1/2, TLR5, and TLR2/6) other than TLR4. Moreover, the effect of gastrointestinal metabolism on the immunomodulatory effects of PYC was investigated.
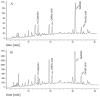
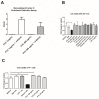
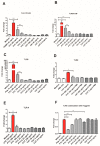
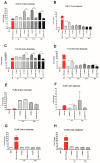
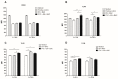
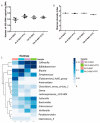
Findings: We showed that intact non-metabolized PYC dose-dependently acts as an agonist of TLR1/2 and TLR2/6 and as a partial agonist of TLR5. PYC on its own does not agonize or antagonize TLR4. However, after the formation of complexes with lipopolysaccharides (LPS), it is a potent activator of TLR4 signaling. Gastrointestinal metabolism of PYC revealed the immunosuppressive potential of the retentate fraction against TLR1/2 and TLR2/6 when compared to the control fraction containing microbiota and enzymes only. The dialyzed fraction containing PYC metabolites revealed the capacity to induce anti-inflammatory IL-10 secretion. Finally, microbially metabolized PYC affected the colonic microbiota composition during in vitro gastrointestinal digestion.
Conclusions: This study showed that gastrointestinal metabolism of PYC reveals its biological activity as a potential inhibitor of TLRs signaling. The results suggest that metabolized PYC acts as a partial agonist of TLR1/2 and TLR2/6 in the presence of the microbiota-derived TLR agonists (retentate fraction) and that it possesses anti-inflammatory potential reflected by the induction of IL-10 from THP-1 macrophages (dialysate fraction).
Keywords: Pycnogenol®; Toll-like receptors; catechin; gastrointestinal metabolism; immunomodulation; metabolites; partial agonist.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Chovanová Z., Muchová J., Sivoňová M., Dvořáková M., Žitňanová I., Waczulíková I., Trebatická J., Škodáček I., Ďuračková Z. Effect of polyphenolic extract, pycnogenol®, on the level of 8-oxoguanine in children suffering from attention deficit/hyperactivity disorder. Free Radic. Res. 2006;40:1003–1010. doi: 10.1080/10715760600824902. - DOI - PubMed
-
- Rohdewald P. Pycnogenol. In: Rice-Evans C.A., Packer L., editors. Flavonoids in Health and Disease. Marcel Dekker, Inc.; New York, NY, USA: 1998.
-
- Mülek M., Seefried L., Genest F., Högger P. Distribution of Constituents and Metabolites of Maritime Pine Bark Extract (Pycnogenol®) into Serum, Blood Cells, and Synovial Fluid of Patients with Severe Osteoarthritis: A Randomized Controlled Trial. Nutrients. 2017;9:443. doi: 10.3390/nu9050443. - DOI - PMC - PubMed
-
- Grimm T., Skrabala R., Chovanová Z., Muchová J., Sumegová K., Liptáková A., Ďuračková Z., Högger P. Single and multiple dose pharmacokinetics of maritime pine bark extract (Pycnogenol) after oral administration to healthy volunteers. BMC Clin. Pharmacol. 2006;6:4. doi: 10.1186/1472-6904-6-4. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

