Multiple QTL underlie milk phenotypes at the CSF2RB locus
- PMID: 30678637
- PMCID: PMC6346582
- DOI: 10.1186/s12711-019-0446-x
Multiple QTL underlie milk phenotypes at the CSF2RB locus
Abstract
Background: Over many years, artificial selection has substantially improved milk production by cows. However, the genes that underlie milk production quantitative trait loci (QTL) remain relatively poorly characterised. Here, we investigate a previously reported QTL located at the CSF2RB locus on chromosome 5, for several milk production phenotypes, to better understand its underlying genetic and molecular causes.
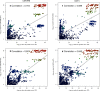
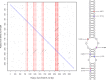
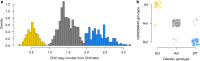
Results: Using a population of 29,350 taurine dairy cows, we conducted association analyses for milk yield and composition traits, and identified highly significant QTL for milk yield, milk fat concentration, and milk protein concentration. Strikingly, protein concentration and milk yield appear to show co-located yet genetically distinct QTL. To attempt to understand the molecular mechanisms that might be mediating these effects, gene expression data were used to investigate eQTL for 11 genes in the broader interval. This analysis highlighted genetic impacts on CSF2RB and NCF4 expression that share similar association signatures to those observed for lactation QTL, strongly implicating one or both of these genes as responsible for these effects. Using the same gene expression dataset representing 357 lactating cows, we also identified 38 novel RNA editing sites in the 3' UTR of CSF2RB transcripts. The extent to which two of these sites were edited also appears to be genetically co-regulated with lactation QTL, highlighting a further layer of regulatory complexity that involves the CSF2RB gene.
Conclusions: This locus presents a diversity of molecular and lactation QTL, likely representing multiple overlapping effects that, at a minimum, highlight the CSF2RB gene as having a causal role in these processes.
Figures





References
-
- Raven LA, Cocks BG, Kemper KE, Chamberlain AJ, Vander Jagt CJ, Goddard ME, et al. Targeted imputation of sequence variants and gene expression profiling identifies twelve candidate genes associated with lactation volume, composition and calving interval in dairy cattle. Mamm Genome. 2016;27:81–97. doi: 10.1007/s00335-015-9613-8. - DOI - PubMed
-
- Pausch H, Emmerling R, Gredler-Grandl B, Fries R, Daetwyler HD, Goddard ME. Meta-analysis of sequence-based association studies across three cattle breeds reveals 25 QTL for fat and protein percentages in milk at nucleotide resolution. BMC Genomics. 2017;18:853. doi: 10.1186/s12864-017-4263-8. - DOI - PMC - PubMed
-
- Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, et al. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002;12:222–231. doi: 10.1101/gr.224202. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

