Genome-wide signatures of local adaptation among seven stoneflies species along a nationwide latitudinal gradient in Japan
- PMID: 30678640
- PMCID: PMC6346529
- DOI: 10.1186/s12864-019-5453-3
Genome-wide signatures of local adaptation among seven stoneflies species along a nationwide latitudinal gradient in Japan
Abstract
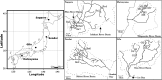
Background: Environmental heterogeneity continuously produces a selective pressure that results in genomic variation among organisms; understanding this relationship remains a challenge in evolutionary biology. Here, we evaluated the degree of genome-environmental association of seven stonefly species across a wide geographic area in Japan and additionally identified putative environmental drivers and their effect on co-existing multiple stonefly species. Double-digest restriction-associated DNA (ddRAD) libraries were independently sequenced for 219 individuals from 23 sites across four geographical regions along a nationwide latitudinal gradient in Japan.
Results: A total of 4251 candidate single nucleotide polymorphisms (SNPs) strongly associated with local adaptation were discovered using Latent mixed models; of these, 294 SNPs showed strong correlation with environmental variables, specifically precipitation and altitude, using distance-based redundancy analysis. Genome-genome comparison among the seven species revealed a high sequence similarity of candidate SNPs within a geographical region, suggesting the occurrence of a parallel evolution process.
Conclusions: Our results revealed genomic signatures of local adaptation and their influence on multiple, co-occurring species. These results can be potentially applied for future studies on river management and climatic stressor impacts.
Keywords: Adaptation; Environmental associations; Landscape genetics; ddRAD.
Conflict of interest statement
Ethics approval and consent to participate
The field sampling was conducted without the necessity of any specific permit.
Consent for publication
No applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Differences in protein expression among five species of stream stonefly (Plecoptera) along a latitudinal gradient in Japan.Arch Insect Biochem Physiol. 2017 Nov;96(3). doi: 10.1002/arch.21422. Epub 2017 Sep 19. Arch Insect Biochem Physiol. 2017. PMID: 28925517
-
Genomic signatures of parallel alpine adaptation in recently evolved flightless insects.Mol Ecol. 2021 Dec;30(24):6677-6686. doi: 10.1111/mec.16204. Epub 2021 Oct 11. Mol Ecol. 2021. PMID: 34592029
-
Signatures of polygenic adaptation associated with climate across the range of a threatened fish species with high genetic connectivity.Mol Ecol. 2017 Nov;26(22):6253-6269. doi: 10.1111/mec.14368. Epub 2017 Nov 8. Mol Ecol. 2017. PMID: 28977721
-
The search for loci under selection: trends, biases and progress.Mol Ecol. 2018 Mar;27(6):1342-1356. doi: 10.1111/mec.14549. Epub 2018 Mar 30. Mol Ecol. 2018. PMID: 29524276 Review.
-
Genomic resources and their influence on the detection of the signal of positive selection in genome scans.Mol Ecol. 2016 Jan;25(1):170-84. doi: 10.1111/mec.13468. Epub 2015 Dec 17. Mol Ecol. 2016. PMID: 26562485 Review.
Cited by
-
ddRAD-Seq reveals evolutionary insights into population differentiation and the cryptic phylogeography of Hyporhamphus intermedius in Mainland China.Ecol Evol. 2022 Jul 4;12(7):e9053. doi: 10.1002/ece3.9053. eCollection 2022 Jul. Ecol Evol. 2022. PMID: 35813915 Free PMC article.
-
Wolbachia-driven selective sweep in a range expanding insect species.BMC Ecol Evol. 2021 Sep 25;21(1):181. doi: 10.1186/s12862-021-01906-6. BMC Ecol Evol. 2021. PMID: 34563127 Free PMC article.
-
Genome-wide detection of Wolbachia in natural Aedes aegypti populations using ddRAD-Seq.Front Cell Infect Microbiol. 2023 Dec 14;13:1252656. doi: 10.3389/fcimb.2023.1252656. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38162582 Free PMC article.
-
Spatial and phylogenetic structure of Alpine stonefly assemblages across seven habitats using DNA-species.Oecologia. 2023 Feb;201(2):513-524. doi: 10.1007/s00442-023-05321-0. Epub 2023 Jan 21. Oecologia. 2023. PMID: 36680607
-
Population Genomics of an Anadromous Hilsa Shad Tenualosa ilisha Species across Its Diverse Migratory Habitats: Discrimination by Fine-Scale Local Adaptation.Genes (Basel). 2019 Dec 30;11(1):46. doi: 10.3390/genes11010046. Genes (Basel). 2019. PMID: 31905942 Free PMC article.
References
-
- Holt RD, Gaines MS. Analysis of adaptation in heterogeneous landscapes: implications for the evolution of fundamental niches. Evol Ecol. 1992;6:433–447. doi: 10.1007/BF02270702. - DOI
-
- Hendrick PW. Genetic polymorphism in heterogeneous environments: the age of genomics. Ann Rev Ecol Evol Syst. 2006;37:67–93. doi: 10.1146/annurev.ecolsys.37.091305.110132. - DOI
-
- Holderegger R, Kamm R, Gugerli F. Adaptive vs. neutral genetic diversity: implications for landscape genetics. Landsc Ecol. 2006;21:797–807. doi: 10.1007/s10980-005-5245-9. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

