MoChlo: A Versatile, Modular Cloning Toolbox for Chloroplast Biotechnology
- PMID: 30679266
- PMCID: PMC6393787
- DOI: 10.1104/pp.18.01220
MoChlo: A Versatile, Modular Cloning Toolbox for Chloroplast Biotechnology
Abstract
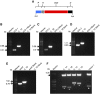
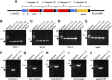
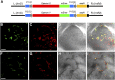
Plant synthetic biology is a rapidly evolving field with new tools constantly emerging to drive innovation. Of particular interest is the application of synthetic biology to chloroplast biotechnology to generate plants capable of producing new metabolites, vaccines, biofuels, and high-value chemicals. Progress made in the assembly of large DNA molecules, composing multiple transcriptional units, has significantly aided in the ability to rapidly construct novel vectors for genetic engineering. In particular, Golden Gate assembly has provided a facile molecular tool for standardized assembly of synthetic genetic elements into larger DNA constructs. In this work, a complete modular chloroplast cloning system, MoChlo, was developed and validated for fast and flexible chloroplast engineering in plants. A library of 128 standardized chloroplast-specific parts (47 promoters, 38 5' untranslated regions [5'UTRs], nine promoter:5'UTR fusions, 10 3'UTRs, 14 genes of interest, and 10 chloroplast-specific destination vectors) were mined from the literature and modified for use in MoChlo assembly, along with chloroplast-specific destination vectors. The strategy was validated by assembling synthetic operons of various sizes and determining the efficiency of assembly. This method was successfully used to generate chloroplast transformation vectors containing up to seven transcriptional units in a single vector (∼10.6-kb synthetic operon). To enable researchers with limited resources to engage in chloroplast biotechnology, and to accelerate progress in the field, the entire kit, as described, is available through Addgene at minimal cost. Thus, the MoChlo kit represents a valuable tool for fast and flexible design of heterologous metabolic pathways for plastid metabolic engineering.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures





References
-
- Baecker JJ, Sneddon JC, Hollingsworth MJ (2009) Efficient translation in chloroplasts requires element(s) upstream of the putative ribosome binding site from atpI. Am J Bot 96: 627–636 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

