A chromosome-scale assembly of the axolotl genome
- PMID: 30679309
- PMCID: PMC6360810
- DOI: 10.1101/gr.241901.118
A chromosome-scale assembly of the axolotl genome
Abstract
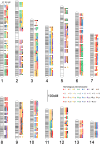
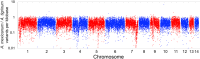
The axolotl (Ambystoma mexicanum) provides critical models for studying regeneration, evolution, and development. However, its large genome (∼32 Gb) presents a formidable barrier to genetic analyses. Recent efforts have yielded genome assemblies consisting of thousands of unordered scaffolds that resolve gene structures, but do not yet permit large-scale analyses of genome structure and function. We adapted an established mapping approach to leverage dense SNP typing information and for the first time assemble the axolotl genome into 14 chromosomes. Moreover, we used fluorescence in situ hybridization to verify the structure of these 14 scaffolds and assign each to its corresponding physical chromosome. This new assembly covers 27.3 Gb and encompasses 94% of annotated gene models on chromosomal scaffolds. We show the assembly's utility by resolving genome-wide orthologies between the axolotl and other vertebrates, identifying the footprints of historical introgression events that occurred during the development of axolotl genetic stocks, and precisely mapping several phenotypes including a large deletion underlying the cardiac mutant. This chromosome-scale assembly will greatly facilitate studies of the axolotl in biological research.
© 2019 Smith et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Armstrong JB, Duhon ST, Malacinski GM. 1989. Raising the axolotl in captivity. In Developmental biology of the axolotl (ed. Armstrong JB, Malacinski GM), pp. 220–227. Oxford University Press, New York.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
