Effects of the long-term storage of human fecal microbiota samples collected in RNAlater
- PMID: 30679604
- PMCID: PMC6345939
- DOI: 10.1038/s41598-018-36953-5
Effects of the long-term storage of human fecal microbiota samples collected in RNAlater
Abstract
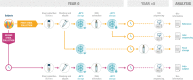
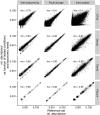
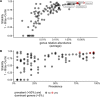
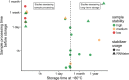
The adequate storage of fecal samples from clinical trials is crucial if analyses are to be performed later and in long-term studies. However, it is unknown whether the composition of the microbiota is preserved during long-term stool storage (>1 year). We therefore evaluated the influence of long-term storage on the microbiota composition of human stool samples collected in RNAlater and stored for approximately five years at -80 °C. We compared storage effects on stool samples from 24 subjects with the effects of technical variation due to different sequencing runs and biological variation (intra- and inter-subject), in another 101 subjects, based on alpha-diversity, beta-diversity and taxonomic composition. We also evaluated the impact of initial alpha-diversity and fecal microbiota composition on beta-diversity instability upon storage. Overall, long-term stool storage at -80 °C had only limited effects on the microbiota composition of human feces. The magnitude of changes in alpha- and beta- diversity and taxonomic composition after long-term storage was similar to inter-sequencing variation and smaller than biological variation (both intra- and inter-subject). The likelihood of fecal samples being affected by long-term storage correlated with the initial relative abundance of some genera and tend to be affected by initial taxonomic richness.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

