The evolution of transcriptional repressors in the Notch signaling pathway: a computational analysis
- PMID: 30679936
- PMCID: PMC6337844
- DOI: 10.1186/s41065-019-0081-0
The evolution of transcriptional repressors in the Notch signaling pathway: a computational analysis
Abstract
Background: The Notch signaling pathway governs the specification of different cell types in flies, nematodes and vertebrates alike. Principal components of the pathway that activate Notch target genes are highly conserved throughout the animal kingdom. Despite the impact on development and disease, repression mechanisms are less well studied. Repressors are known from arthropods and vertebrates that differ strikingly by mode of action: whereas Drosophila Hairless assembles repressor complexes with CSL transcription factors, competition between activator and repressors occurs in vertebrates (for example SHARP/MINT and KyoT2). This divergence raises questions on the evolution: Are there common ancestors throughout the animal kingdom?
Results: Available genome databases representing all animal clades were searched for homologues of Hairless, SHARP and KyoT2. The most distant species with convincing Hairless orthologs belong to Myriapoda, indicating its emergence after the Mandibulata-Chelicarata radiation about 500 million years ago. SHARP shares motifs with SPEN and SPENITO proteins, present throughout the animal kingdom. The CSL interacting domain of SHARP, however, is specific to vertebrates separated by roughly 600 million years of evolution. KyoT2 bears a C-terminal CSL interaction domain (CID), present only in placental mammals but highly diverged already in marsupials, suggesting introduction roughly 100 million years ago. Based on the LIM-domains that characterize KyoT2, homologues can be found in Drosophila melanogaster (Limpet) and Hydra vulgaris (Prickle 3 like). These lack the CID of KyoT2, however, contain a PET and additional LIM domains. Conservation of intron/exon boundaries underscores the phylogenetic relationship between KyoT2, Limpet and Prickle. Most strikingly, Limpet and Prickle proteins carry a tetra-peptide motif resembling that of several CSL interactors. Overall, KyoT2 may have evolved from prickle and Limpet to a Notch repressor in mammals.
Conclusions: Notch repressors appear to be specific to either chordates or arthropods. Orthologues of experimentally validated repressors were not found outside the phylogenetic group they have been originally identified. However, the data provide a hypothesis on the evolution of mammalian KyoT2 from Prickle like ancestors. The finding of a potential CSL interacting domain in Prickle homologues points to a novel, very ancestral CSL interactor present in the entire animal kingdom.
Keywords: Animal kingdom; Evolution; Gene annotation; Hairless; KyoT2; Limpet; Notch repressors; Prickle; SHARP.
Conflict of interest statement
Not applicable.Not applicable.The author declares that there are no competing financial, personal, or professional interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



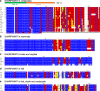



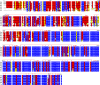



References
-
- Dexter JS. The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am Nat. 1914;48:712–758. doi: 10.1086/279446. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

