Identification of genes associated with ricinoleic acid accumulation in Hiptage benghalensis via transcriptome analysis
- PMID: 30679955
- PMCID: PMC6340187
- DOI: 10.1186/s13068-019-1358-2
Identification of genes associated with ricinoleic acid accumulation in Hiptage benghalensis via transcriptome analysis
Abstract
Background: Ricinoleic acid is a high-value hydroxy fatty acid with broad industrial applications. Hiptage benghalensis seed oil contains a high amount of ricinoleic acid (~ 80%) and represents an emerging source of this unusual fatty acid. However, the mechanism of ricinoleic acid accumulation in H. benghalensis is yet to be explored at the molecular level, which hampers the exploration of its potential in ricinoleic acid production.
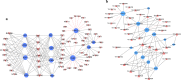
Results: To explore the molecular mechanism of ricinoleic acid biosynthesis and regulation, H. benghalensis seeds were harvested at five developing stages (13, 16, 19, 22, and 25 days after pollination) for lipid analysis. The results revealed that the rapid accumulation of ricinoleic acid occurred at the early-mid-seed development stages (16-22 days after pollination). Subsequently, the gene transcription profiles of the developing seeds were characterized via a comprehensive transcriptome analysis with second-generation sequencing and single-molecule real-time sequencing. Differential expression patterns were identified in 12,555 transcripts, including 71 enzymes in lipid metabolic pathways, 246 putative transcription factors (TFs) and 124 long noncoding RNAs (lncRNAs). Twelve genes involved in diverse lipid metabolism pathways, including fatty acid biosynthesis and modification (hydroxylation), lipid traffic, triacylglycerol assembly, acyl editing and oil-body formation, displayed high expression levels and consistent expression patterns with ricinoleic acid accumulation in the developing seeds, suggesting their primary roles in ricinoleic acid production. Subsequent co-expression network analysis identified 57 TFs and 35 lncRNAs, which are putatively involved in the regulation of ricinoleic acid biosynthesis. The transcriptome data were further validated by analyzing the expression profiles of key enzyme-encoding genes, TFs and lncRNAs with quantitative real-time PCR. Finally, a network of genes associated with ricinoleic acid accumulation in H. benghalensis was established.
Conclusions: This study was the first step toward the understating of the molecular mechanisms of ricinoleic acid biosynthesis and oil accumulation in H. benghalensis seeds and identified a pool of novel genes regulating ricinoleic acid accumulation. The results set a foundation for developing H. benghalensis into a novel ricinoleic acid feedstock at the transcriptomic level and provided valuable candidate genes for improving ricinoleic acid production in other plants.
Keywords: Co-expression network analysis; Hiptage benghalensis; Industrial oils; Lipid biosynthesis; Long noncoding RNA; Oilseed; RNA-Seq; Ricinoleic acid; Transcription factor; Transcriptomics.
Figures





References
-
- McKeon TA. Castor (Ricinus communis L.) In: McKeon T, Hayes DG, Hildebrand DF, Weselake RJ, editors. Industrial oil crops. New York: Elsevier/AOCS Press; 2016. pp. 75–112.
-
- Severino LS, Auld DL, Baldanzi M, Cândido MJ, Chen G, Crosby W, et al. A review on the challenges for increased production of castor. Agron J. 2012;104:853–880. doi: 10.2134/agronj2011.0210. - DOI
-
- Castor oil world, Global demand and supply of castor oil. https://www.castoroilworld.com/statistics-market-demand-future-trend/. Accessed 04 Oct 2018.
LinkOut - more resources
Full Text Sources

