Interaction of germline variants in a family with a history of early-onset clear cell renal cell carcinoma
- PMID: 30680959
- PMCID: PMC6418363
- DOI: 10.1002/mgg3.556
Interaction of germline variants in a family with a history of early-onset clear cell renal cell carcinoma
Abstract
Background: Identification of genetic factors causing predisposition to renal cell carcinoma has helped improve screening, early detection, and patient survival.
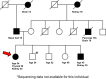
Methods: We report the characterization of a proband with renal and thyroid cancers and a family history of renal and other cancers by whole-exome sequencing (WES), coupled with WES analysis of germline DNA from additional affected and unaffected family members.
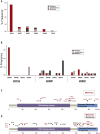
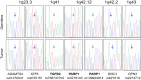
Results: This work identified multiple predicted protein-damaging variants relevant to the pattern of inherited cancer risk. Among these, the proband and an affected brother each had a heterozygous Ala45Thr variant in SDHA, a component of the succinate dehydrogenase (SDH) complex. SDH defects are associated with mitochondrial disorders and risk for various cancers; immunochemical analysis indicated loss of SDHB protein expression in the patient's tumor, compatible with SDH deficiency. Integrated analysis of public databases and structural predictions indicated that the two affected individuals also had additional variants in genes including TGFB2, TRAP1, PARP1, and EGF, each potentially relevant to cancer risk alone or in conjunction with the SDHA variant. In addition, allelic imbalances of PARP1 and TGFB2 were detected in the tumor of the proband.
Conclusion: Together, these data suggest the possibility of risk associated with interaction of two or more variants.
Keywords: cancer risk; germline; renal cell carcinoma; succinate dehydrogenase complex; variant interaction; variants of uncertain significance.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
None declared.
Figures


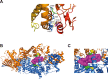
References
-
- Bannon, A. E. , Kent, J. , Forquer, I. , Town, A. , Klug, L. R. , McCann, K. , … Corless, C. (2017). Biochemical, molecular, and clinical characterization of succinate dehydrogenase subunit A variants of unknown significance. Clinical Cancer Research, 23(21), 6733–6743. 10.1158/1078-0432.CCR-17-1397 - DOI - PMC - PubMed
-
- Belinsky, M. G. , Rink, L. , Flieder, D. B. , Jahromi, M. S. , Schiffman, J. D. , Godwin, A. K. , & Mehren, M. (2013). Overexpression of insulin‐like growth factor 1 receptor and frequent mutational inactivation of SDHA in wild‐type SDHB‐negative gastrointestinal stromal tumors. Genes, Chromosomes & Cancer, 52(2), 214–224. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

