Requirements for RNA polymerase II preinitiation complex formation in vivo
- PMID: 30681409
- PMCID: PMC6366898
- DOI: 10.7554/eLife.43654
Requirements for RNA polymerase II preinitiation complex formation in vivo
Abstract
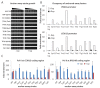
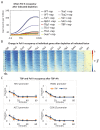
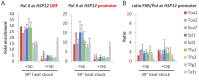
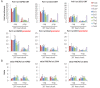
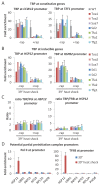
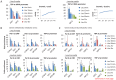
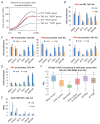
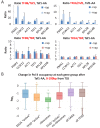
Transcription by RNA polymerase II requires assembly of a preinitiation complex (PIC) composed of general transcription factors (GTFs) bound at the promoter. In vitro, some GTFs are essential for transcription, whereas others are not required under certain conditions. PICs are stable in the absence of nucleotide triphosphates, and subsets of GTFs can form partial PICs. By depleting individual GTFs in yeast cells, we show that all GTFs are essential for TBP binding and transcription, suggesting that partial PICs do not exist at appreciable levels in vivo. Depletion of FACT, a histone chaperone that travels with elongating Pol II, strongly reduces PIC formation and transcription. In contrast, TBP-associated factors (TAFs) contribute to transcription of most genes, but TAF-independent transcription occurs at substantial levels, preferentially at promoters containing TATA elements. PICs are absent in cells deprived of uracil, and presumably UTP, suggesting that transcriptionally inactive PICs are removed from promoters in vivo.
Keywords: RNA polymerase II; S. cerevisiae; TATA-binding protein; TBP-associated factors (TAFs); chromosomes; gene expression; general transcription factors; genetics; genomics; preinitiation complex; transcription.
© 2019, Petrenko et al.
Conflict of interest statement
NP, YJ, LD, KW No competing interests declared, KS Senior editor, eLife
Figures




Similar articles
-
Comparison of transcriptional initiation by RNA polymerase II across eukaryotic species.Elife. 2021 Sep 13;10:e67964. doi: 10.7554/eLife.67964. Elife. 2021. PMID: 34515029 Free PMC article.
-
Enhancement of TBP binding by activators and general transcription factors.Nature. 1999 Jun 10;399(6736):605-9. doi: 10.1038/21232. Nature. 1999. PMID: 10376604
-
Independent RNA polymerase II preinitiation complex dynamics and nucleosome turnover at promoter sites in vivo.Genome Res. 2014 Jan;24(1):117-24. doi: 10.1101/gr.157792.113. Epub 2013 Dec 2. Genome Res. 2014. PMID: 24298073 Free PMC article.
-
Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription.Gene. 2003 Oct 2;315:1-13. doi: 10.1016/s0378-1119(03)00714-5. Gene. 2003. PMID: 14557059 Review.
-
The general transcription machinery and general cofactors.Crit Rev Biochem Mol Biol. 2006 May-Jun;41(3):105-78. doi: 10.1080/10409230600648736. Crit Rev Biochem Mol Biol. 2006. PMID: 16858867 Review.
Cited by
-
The TFIIH complex is required to establish and maintain mitotic chromosome structure.Elife. 2022 Mar 16;11:e75475. doi: 10.7554/eLife.75475. Elife. 2022. PMID: 35293859 Free PMC article.
-
Centromeric Transcription: A Conserved Swiss-Army Knife.Genes (Basel). 2020 Aug 9;11(8):911. doi: 10.3390/genes11080911. Genes (Basel). 2020. PMID: 32784923 Free PMC article. Review.
-
Kin28 depletion increases association of TFIID subunits Taf1 and Taf4 with promoters in Saccharomyces cerevisiae.Nucleic Acids Res. 2020 May 7;48(8):4244-4255. doi: 10.1093/nar/gkaa165. Nucleic Acids Res. 2020. PMID: 32182349 Free PMC article.
-
Genome-scale chromatin binding dynamics of RNA Polymerase II general transcription machinery components.EMBO J. 2024 May;43(9):1799-1821. doi: 10.1038/s44318-024-00089-2. Epub 2024 Apr 2. EMBO J. 2024. PMID: 38565951 Free PMC article.
-
TBP facilitates RNA Polymerase I transcription following mitosis.RNA Biol. 2024 Jan;21(1):42-51. doi: 10.1080/15476286.2024.2375097. Epub 2024 Jul 3. RNA Biol. 2024. PMID: 38958280 Free PMC article.
References
-
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Molecular Biology of the Cell. 2008;19:352–367. doi: 10.1091/mbc.e07-08-0779. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

