Role of CsrA in stress responses and metabolism important for Salmonella virulence revealed by integrated transcriptomics
- PMID: 30682134
- PMCID: PMC6347204
- DOI: 10.1371/journal.pone.0211430
Role of CsrA in stress responses and metabolism important for Salmonella virulence revealed by integrated transcriptomics
Abstract
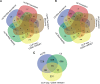
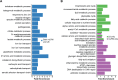
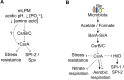
To cause infection, Salmonella must survive and replicate in host niches that present dramatically different environmental conditions. This requires a flexible metabolism and physiology, responsive to conditions of the local milieu. The sequence specific RNA binding protein CsrA serves as a global regulator that governs gene expression required for pathogenicity, metabolism, biofilm formation, and motility in response to nutritional conditions. Its activity is determined by two noncoding small RNAs (sRNA), CsrB and CsrC, which sequester and antagonize this protein. Here, we used ribosome profiling and RNA-seq analysis to comprehensively examine the effects of CsrA on mRNA occupancy with ribosomes, a measure of translation, transcript stability, and the steady state levels of transcripts under in vitro SPI-1 inducing conditions, to simulate growth in the intestinal lumen, and under in vitro SPI-2-inducing conditions, to simulate growth in the Salmonella containing vacuole (SCV) of the macrophage. Our findings uncovered new roles for CsrA in controlling the expression of structural and regulatory genes involved in stress responses, metabolism, and virulence systems required for infection. We observed substantial variation in the CsrA regulon under the two growth conditions. In addition, CsrB/C sRNA levels were greatly reduced under the simulated intracellular conditions and were responsive to nutritional factors that distinguish the intracellular and luminal environments. Altogether, our results reveal CsrA to be a flexible regulator, which is inferred to be intimately involved in maintaining the distinct gene expression patterns associated with growth in the intestine and the macrophage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLOS Med. 2015;12: e1001921 10.1371/journal.pmed.1001921 - DOI - PMC - PubMed
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382: 209–22. 10.1016/S0140-6736(13)60844-2 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

