Influenza-induced immune suppression to methicillin-resistant Staphylococcus aureus is mediated by TLR9
- PMID: 30682165
- PMCID: PMC6364947
- DOI: 10.1371/journal.ppat.1007560
Influenza-induced immune suppression to methicillin-resistant Staphylococcus aureus is mediated by TLR9
Abstract
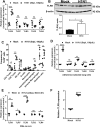
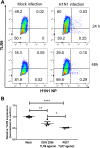
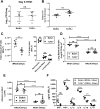
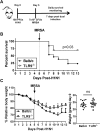
Bacterial lung infections, particularly with methicillin-resistant Staphylococcus aureus (MRSA), increase mortality following influenza infection, but the mechanisms remain unclear. Here we show that expression of TLR9, a microbial DNA sensor, is increased in murine lung macrophages, dendritic cells, CD8+ T cells and epithelial cells post-influenza infection. TLR9-/- mice did not show differences in handling influenza nor MRSA infection alone. However, TLR9-/- mice have improved survival and bacterial clearance in the lung post-influenza and MRSA dual infection, with no difference in viral load during dual infection. We demonstrate that TLR9 is upregulated on macrophages even when they are not themselves infected, suggesting that TLR9 upregulation is related to soluble mediators. We rule out a role for elevations in interferon-γ (IFNγ) in mediating the beneficial MRSA clearance in TLR9-/- mice. While macrophages from WT and TLR9-/- mice show similar phagocytosis and bacterial killing to MRSA alone, following influenza infection, there is a marked upregulation of scavenger receptor A and MRSA phagocytosis as well as inducible nitric oxide synthase (Inos) and improved bacterial killing that is specific to TLR9-deficient cells. Bone marrow transplant chimera experiments and in vitro experiments using TLR9 antagonists suggest TLR9 expression on non-hematopoietic cells, rather than the macrophages themselves, is important for regulating myeloid cell function. Interestingly, improved bacterial clearance post-dual infection was restricted to MRSA, as there was no difference in the clearance of Streptococcus pneumoniae. Taken together these data show a surprising inhibitory role for TLR9 signaling in mediating clearance of MRSA that manifests following influenza infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Comment in
-
Influenza and Staphylococcus aureus Coinfection: TLR9 at Play.Trends Microbiol. 2019 May;27(5):383-384. doi: 10.1016/j.tim.2019.02.006. Epub 2019 Mar 11. Trends Microbiol. 2019. PMID: 30871857
References
-
- Dunning J, Blankley S, Hoang LT, Cox M, Graham CM, James PL, et al. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nature immunology. 2018;19(6):625–35. Epub 2018/05/20. 10.1038/s41590-018-0111-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

