Multiview Cluster Analysis Identifies Variable Corticosteroid Response Phenotypes in Severe Asthma
- PMID: 30682261
- PMCID: PMC6543720
- DOI: 10.1164/rccm.201808-1543OC
Multiview Cluster Analysis Identifies Variable Corticosteroid Response Phenotypes in Severe Asthma
Erratum in
-
Erratum: Multiview Cluster Analysis Identifies Variable Corticosteroid Response Phenotypes in Severe Asthma.Am J Respir Crit Care Med. 2019 Aug 1;200(3):400. doi: 10.1164/rccm.v200erratum2. Am J Respir Crit Care Med. 2019. PMID: 31368796 Free PMC article. No abstract available.
Abstract
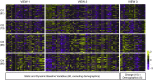
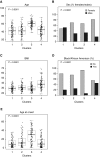
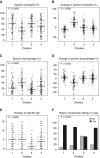
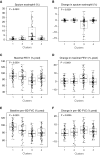
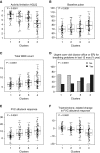
Rationale: Corticosteroids (CSs) are the most effective asthma therapy, but responses are heterogeneous and systemic CSs lead to long-term side effects. Therefore, an improved understanding of the contributing factors in CS responses could enhance precision management. Although several factors have been associated with CS responsiveness, no integrated/cluster approach has yet been undertaken to identify differential CS responses. Objectives: To identify asthma subphenotypes with differential responses to CS treatment using an unsupervised multiview learning approach. Methods: Multiple-kernel k-means clustering was applied to 100 clinical, physiological, inflammatory, and demographic variables from 346 adult participants with asthma in the Severe Asthma Research Program with paired (before and 2-3 weeks after triamcinolone administration) sputum data. Machine-learning techniques were used to select the top baseline variables that predicted cluster assignment for a new patient. Measurements and Main Results: Multiple-kernel clustering revealed four clusters of individuals with asthma and different CS responses. Clusters 1 and 2 consisted of young, modestly CS-responsive individuals with allergic asthma and relatively normal lung function, separated by contrasting sputum neutrophil and macrophage percentages after CS treatment. The subjects in cluster 3 had late-onset asthma and low lung function, high baseline eosinophilia, and the greatest CS responsiveness. Cluster 4 consisted primarily of young, obese females with severe airflow limitation, little eosinophilic inflammation, and the least CS responsiveness. The top 12 baseline variables were identified, and the clusters were validated using an independent Severe Asthma Research Program test set. Conclusions: Our machine learning-based approaches provide new insights into the mechanisms of CS responsiveness in asthma, with the potential to improve disease treatment.
Keywords: asthma phenotype; corticosteroids; eosinophils; severe asthma.
Figures
Comment in
-
Predicting Response to Triamcinolone in Severe Asthma by Machine Learning. Solving the Enigma.Am J Respir Crit Care Med. 2019 Jun 1;199(11):1299-1300. doi: 10.1164/rccm.201902-0320ED. Am J Respir Crit Care Med. 2019. PMID: 30789751 Free PMC article. No abstract available.
References
-
- Wenzel SE. Asthma: defining of the persistent adult phenotypes. Lancet. 2006;368:804–813. - PubMed
-
- Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. - PubMed
-
- Buchman AL. Side effects of corticosteroid therapy. J Clin Gastroenterol. 2001;33:289–294. - PubMed
-
- Dahl R. Systemic side effects of inhaled corticosteroids in patients with asthma. Respir Med. 2006;100:1307–1317. - PubMed
-
- Djukanović R, Wilson JW, Britten KM, Wilson SJ, Walls AF, Roche WR, et al. Effect of an inhaled corticosteroid on airway inflammation and symptoms in asthma. Am Rev Respir Dis. 1992;145:669–674. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U10 HL109257/HL/NHLBI NIH HHS/United States
- UL1 TR000427/TR/NCATS NIH HHS/United States
- U10 HL109250/HL/NHLBI NIH HHS/United States
- U10 HL109164/HL/NHLBI NIH HHS/United States
- U10 HL109172/HL/NHLBI NIH HHS/United States
- U10 HL064313/HL/NHLBI NIH HHS/United States
- R01 HL122531/HL/NHLBI NIH HHS/United States
- P30 DA035778/DA/NIDA NIH HHS/United States
- R01 GM114311/GM/NIGMS NIH HHS/United States
- UL1 TR000454/TR/NCATS NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- UL1 TR002373/TR/NCATS NIH HHS/United States
- U10 HL109086/HL/NHLBI NIH HHS/United States
- U10 HL109168/HL/NHLBI NIH HHS/United States
- UL1 TR001102/TR/NCATS NIH HHS/United States
- U10 HL109152/HL/NHLBI NIH HHS/United States
- U10 HL109146/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

