Evolutionary Analysis of GH3 Genes in Six Oryza Species/Subspecies and Their Expression under Salinity Stress in Oryza sativa ssp. japonica
- PMID: 30682815
- PMCID: PMC6409606
- DOI: 10.3390/plants8020030
Evolutionary Analysis of GH3 Genes in Six Oryza Species/Subspecies and Their Expression under Salinity Stress in Oryza sativa ssp. japonica
Abstract
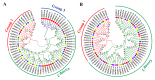
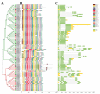
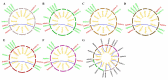
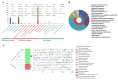
Glycoside Hydrolase 3 (GH3), a member of the Auxin-responsive gene family, is involved in plant growth, the plant developmental process, and various stress responses. The GH3 gene family has been well-studied in Arabidopsis thaliana and Zea mays. However, the evolution of the GH3 gene family in Oryza species remains unknown and the function of the GH3 gene family in Oryza sativa is not well-documented. Here, a systematic analysis was performed in six Oryza species/subspecies, including four wild rice species and two cultivated rice subspecies. A total of 13, 13, 13, 13, 12, and 12 members were identified in O. sativa ssp. japonica, O. sativa ssp. indica, Oryza rufipogon, Oryza nivara, Oryza punctata, and Oryza glumaepatula, respectively. Gene duplication events, structural features, conserved motifs, a phylogenetic analysis, chromosome locations, and Ka/Ks ratios of this important family were found to be strictly conservative across these six Oryza species/subspecies, suggesting that the expansion of the GH3 gene family in Oryza species might be attributed to duplication events, and this expansion could occur in the common ancestor of Oryza species, even in common ancestor of rice tribe (Oryzeae) (23.07~31.01 Mya). The RNA-seq results of different tissues displayed that OsGH3 genes had significantly different expression profiles. Remarkably, the qRT-PCR result after NaCl treatment indicated that the majority of OsGH3 genes play important roles in salinity stress, especially OsGH3-2 and OsGH3-8. This study provides important insights into the evolution of the GH3 gene family in Oryza species and will assist with further investigation of OsGH3 genes' functions under salinity stress.
Keywords: GH3, salinity stress; Oryza species; RNA-seq; gene duplication; qRT-PCR; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Staswick P.E., Tiryaki I., Rowe M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on Jasmonic, Salicylic, and Indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

