Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy
- PMID: 30682877
- PMCID: PMC6384606
- DOI: 10.3390/molecules24030429
Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy
Abstract
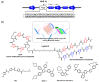
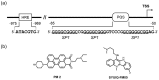
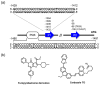
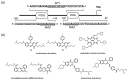
A G-quadruplex (G4) is a well-known nucleic acid secondary structure comprising guanine-rich sequences, and has profound implications for various pharmacological and biological events, including cancers. Therefore, ligands interacting with G4s have attracted great attention as potential anticancer therapies or in molecular probe applications. To date, a large variety of DNA/RNA G4 ligands have been developed by a number of laboratories. As protein-targeting drugs face similar situations, G-quadruplex-interacting drugs displayed low selectivity to the targeted G-quadruplex structure. This low selectivity could cause unexpected effects that are usually reasons to halt the drug development process. In this review, we address the recent research on synthetic G4 DNA-interacting ligands that allow targeting of selected G4s as an approach toward the discovery of highly effective anticancer drugs.
Keywords: G-quadruplex; cancer therapy; oncogenes; selective ligands; telomere.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

