REM sleep's unique associations with corticosterone regulation, apoptotic pathways, and behavior in chronic stress in mice
- PMID: 30683720
- PMCID: PMC6377491
- DOI: 10.1073/pnas.1816456116
REM sleep's unique associations with corticosterone regulation, apoptotic pathways, and behavior in chronic stress in mice
Abstract
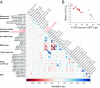
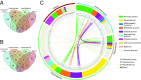
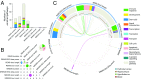
One of sleep's putative functions is mediation of adaptation to waking experiences. Chronic stress is a common waking experience; however, which specific aspect of sleep is most responsive, and how sleep changes relate to behavioral disturbances and molecular correlates remain unknown. We quantified sleep, physical, endocrine, and behavioral variables, as well as the brain and blood transcriptome in mice exposed to 9 weeks of unpredictable chronic mild stress (UCMS). Comparing 46 phenotypic variables revealed that rapid-eye-movement sleep (REMS), corticosterone regulation, and coat state were most responsive to UCMS. REMS theta oscillations were enhanced, whereas delta oscillations in non-REMS were unaffected. Transcripts affected by UCMS in the prefrontal cortex, hippocampus, hypothalamus, and blood were associated with inflammatory and immune responses. A machine-learning approach controlling for unspecific UCMS effects identified transcriptomic predictor sets for REMS parameters that were enriched in 193 pathways, including some involved in stem cells, immune response, and apoptosis and survival. Only three pathways were enriched in predictor sets for non-REMS. Transcriptomic predictor sets for variation in REMS continuity and theta activity shared many pathways with corticosterone regulation, in particular pathways implicated in apoptosis and survival, including mitochondrial apoptotic machinery. Predictor sets for REMS and anhedonia shared pathways involved in oxidative stress, cell proliferation, and apoptosis. These data identify REMS as a core and early element of the response to chronic stress, and identify apoptosis and survival pathways as a putative mechanism by which REMS may mediate the response to stressful waking experiences.
Keywords: EEG theta power; anhedonia; depression; machine learning; transcriptome.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: This study was supported by a Lilly Innovation Fellowship Award (to M.N.) and conducted through an academic–industrial partnership between the Surrey Sleep Research Centre of the University of Surrey and Eli Lilly and Company Ltd. K.A.W., A.P.M., K.M., N.L., and M.N. were full-time employees of Eli Lilly and Company Ltd. at the time of the study. D.-J.D. has received research funds and acted as consultant to Eli Lilly and other pharmaceutical companies. R.W.-S. has received research funding from Eli Lilly.
Figures



References
-
- Sanford LD, Suchecki D, Meerlo P. Stress, arousal, and sleep. Curr Top Behav Neurosci. 2015;25:379–410. - PubMed
-
- Tafet GE, Nemeroff CB. The links between stress and depression: Psychoneuroendocrinological, genetic, and environmental interactions. J Neuropsychiatry Clin Neurosci. 2016;28:77–88. - PubMed
-
- World Health Organization 2008 The Global Burden of Disease: 2004 Update. Available at https://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/. Accessed September 1, 2018.
-
- Vigo D, Thornicroft G, Atun R. Estimating the true global burden of mental illness. Lancet Psychiatry. 2016;3:171–178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

