Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction
- PMID: 30683722
- PMCID: PMC6377443
- DOI: 10.1073/pnas.1812963116
Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction
Abstract
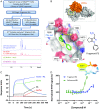
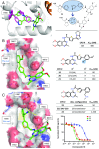
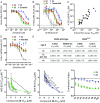
Since the late 1980s, mutations in the RAS genes have been recognized as major oncogenes with a high occurrence rate in human cancers. Such mutations reduce the ability of the small GTPase RAS to hydrolyze GTP, keeping this molecular switch in a constitutively active GTP-bound form that drives, unchecked, oncogenic downstream signaling. One strategy to reduce the levels of active RAS is to target guanine nucleotide exchange factors, which allow RAS to cycle from the inactive GDP-bound state to the active GTP-bound form. Here, we describe the identification of potent and cell-active small-molecule inhibitors which efficiently disrupt the interaction between KRAS and its exchange factor SOS1, a mode of action confirmed by a series of biophysical techniques. The binding sites, mode of action, and selectivity were elucidated using crystal structures of KRASG12C-SOS1, SOS1, and SOS2. By preventing formation of the KRAS-SOS1 complex, these inhibitors block reloading of KRAS with GTP, leading to antiproliferative activity. The final compound 23 (BAY-293) selectively inhibits the KRAS-SOS1 interaction with an IC50 of 21 nM and is a valuable chemical probe for future investigations.
Keywords: RAS; SOS; crystal structure; fragment screen; small-molecule inhibitor.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: R.C.H. B.S., J.S., D.M., A.H., C.M.S., N.D.W., H.B., U.B., J.W., V.B., J.M., K.P., G.S., N.B., K.E., K.G., L.W., F.v.N., and B.B. are or have been employees and stockholders of Bayer AG. J.D.K. and D.W. are employees of Evotec AG. R.C.H., B.S., J.S., D.M., H.B., K.P., N.B., K.E., L.W., F.v.N., and B.B. are coauthors of a patent application.
Figures


References
-
- Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. - PubMed
-
- Santos E, Tronick SR, Aaronson SA, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature. 1982;298:343–347. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

