Primary and Secondary Metabolic Effects of a Key Gene Deletion (Δ YPL062W) in Metabolically Engineered Terpenoid-Producing Saccharomyces cerevisiae
- PMID: 30683746
- PMCID: PMC6585493
- DOI: 10.1128/AEM.01990-18
Primary and Secondary Metabolic Effects of a Key Gene Deletion (Δ YPL062W) in Metabolically Engineered Terpenoid-Producing Saccharomyces cerevisiae
Abstract
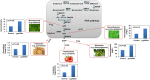
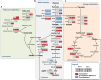
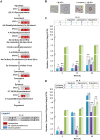
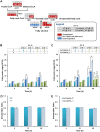
Saccharomyces cerevisiae is an established cell factory for production of terpenoid pharmaceuticals and chemicals. Numerous studies have demonstrated that deletion or overexpression of off-pathway genes in yeast can improve terpenoid production. The deletion of YPL062W in S. cerevisiae, in particular, has benefitted carotenoid production by channeling carbon toward carotenoid precursors acetyl coenzyme A (acetyl-CoA) and mevalonate. The genetic function of YPL062W and the molecular mechanisms for these benefits are unknown. In this study, we systematically examined this gene deletion to uncover the gene function and its molecular mechanism. RNA sequencing (RNA-seq) analysis uncovered that YPL062W deletion upregulated the pyruvate dehydrogenase bypass, the mevalonate pathway, heterologous expression of galactose (GAL) promoter-regulated genes, energy metabolism, and membrane composition synthesis. Bioinformatics analysis and serial promoter deletion assay revealed that YPL062W functions as a core promoter for ALD6 and that the expression level of ALD6 is negatively correlated to terpenoid productivity. We demonstrate that ΔYPL062W increases the production of all major terpenoid classes (C10, C15, C20, C30, and C40). Our study not only elucidated the biological function of YPL062W but also provided a detailed methodology for understanding the mechanistic aspects of strain improvement.IMPORTANCE Although computational and reverse metabolic engineering approaches often lead to improved gene deletion mutants for cell factory engineering, the systems level effects of such gene deletions on the production phenotypes have not been extensively studied. Understanding the genetic and molecular function of such gene alterations on production strains will minimize the risk inherent in the development of large-scale fermentation processes, which is a daunting challenge in the field of industrial biotechnology. Therefore, we established a detailed experimental and systems biology approach to uncover the molecular mechanisms of YPL062W deletion in S. cerevisiae, which is shown to improve the production of all terpenoid classes. This study redefines the genetic function of YPL062W, demonstrates a strong correlation between YPL062W and terpenoid production, and provides a useful modification for the creation of terpenoid production platform strains. Further, this study underscores the benefits of detailed and systematic characterization of the metabolic effects of genetic alterations on engineered biosynthetic factories.
Keywords: ALD6; Saccharomyces cerevisiae; YPL062W; terpenoids.
Copyright © 2019 American Society for Microbiology.
Figures


Similar articles
-
Regulating the metabolic flux of pyruvate dehydrogenase bypass to enhance lipid production in Saccharomyces cerevisiae.Commun Biol. 2024 Oct 26;7(1):1399. doi: 10.1038/s42003-024-07103-7. Commun Biol. 2024. PMID: 39462103 Free PMC article.
-
Metabolic pathway engineering for fatty acid ethyl ester production in Saccharomyces cerevisiae using stable chromosomal integration.J Ind Microbiol Biotechnol. 2015 Mar;42(3):477-86. doi: 10.1007/s10295-014-1540-2. Epub 2014 Nov 25. J Ind Microbiol Biotechnol. 2015. PMID: 25422103
-
In Vivo Validation of In Silico Predicted Metabolic Engineering Strategies in Yeast: Disruption of α-Ketoglutarate Dehydrogenase and Expression of ATP-Citrate Lyase for Terpenoid Production.PLoS One. 2015 Dec 23;10(12):e0144981. doi: 10.1371/journal.pone.0144981. eCollection 2015. PLoS One. 2015. PMID: 26701782 Free PMC article.
-
Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective.Microb Cell Fact. 2017 May 11;16(1):82. doi: 10.1186/s12934-017-0694-9. Microb Cell Fact. 2017. PMID: 28494761 Free PMC article. Review.
-
Multiplexed engineering of cytochrome P450 enzymes for promoting terpenoid synthesis in Saccharomyces cerevisiae cell factories: A review.Biotechnol Adv. 2025 Jul-Aug;81:108560. doi: 10.1016/j.biotechadv.2025.108560. Epub 2025 Mar 9. Biotechnol Adv. 2025. PMID: 40068711 Review.
Cited by
-
Identifying and engineering the ideal microbial terpenoid production host.Appl Microbiol Biotechnol. 2019 Jul;103(14):5501-5516. doi: 10.1007/s00253-019-09892-y. Epub 2019 May 25. Appl Microbiol Biotechnol. 2019. PMID: 31129740 Free PMC article. Review.
-
Comparative transcriptome analyses of different Salvia miltiorrhiza varieties during the accumulation of tanshinones.PeerJ. 2021 Oct 20;9:e12300. doi: 10.7717/peerj.12300. eCollection 2021. PeerJ. 2021. PMID: 34721983 Free PMC article.
-
De novo production of bioactive sesterterpenoid ophiobolins in Saccharomyces cerevisiae cell factories.Microb Cell Fact. 2024 May 6;23(1):129. doi: 10.1186/s12934-024-02406-0. Microb Cell Fact. 2024. PMID: 38711040 Free PMC article.
-
Advances in the Discovery and Engineering of Gene Targets for Carotenoid Biosynthesis in Recombinant Strains.Biomolecules. 2023 Dec 5;13(12):1747. doi: 10.3390/biom13121747. Biomolecules. 2023. PMID: 38136618 Free PMC article. Review.
-
De novo biosynthesis of sakuranetin from glucose by engineered Saccharomyces cerevisiae.Appl Microbiol Biotechnol. 2023 Jun;107(12):3899-3909. doi: 10.1007/s00253-023-12564-7. Epub 2023 May 6. Appl Microbiol Biotechnol. 2023. PMID: 37148336
References
-
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943. doi:10.1038/nature04640. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

