SETD3 is a positive regulator of DNA-damage-induced apoptosis
- PMID: 30683849
- PMCID: PMC6347638
- DOI: 10.1038/s41419-019-1328-4
SETD3 is a positive regulator of DNA-damage-induced apoptosis
Abstract
SETD3 is a member of the protein lysine methyltransferase (PKMT) family, which catalyzes the addition of methyl group to lysine residues. However, the protein network and the signaling pathways in which SETD3 is involved remain largely unexplored. In the current study, we show that SETD3 is a positive regulator of DNA-damage-induced apoptosis in colon cancer cells. Our data indicate that depletion of SETD3 from HCT-116 cells results in a significant inhibition of apoptosis after doxorubicin treatment. Our results imply that the positive regulation is sustained by methylation, though the substrate remains unknown. We present a functional cross-talk between SETD3 and the tumor suppressor p53. SETD3 binds p53 in cells in response to doxorubicin treatment and positively regulates p53 target genes activation under these conditions. Mechanistically, we provide evidence that the presence of SETD3 and its catalytic activity is required for the recruitment of p53 to its target genes. Finally, Kaplan-Meier survival analysis, of two-independent cohorts of colon cancer patients, revealed that low expression of SETD3 is a reliable predictor of poor survival in these patients, which correlates with our findings. Together, our data uncover a new role of the PKMT SETD3 in the regulation of p53-dependent activation of apoptosis in response to DNA damage.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

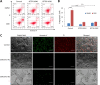
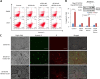


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

