The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma
- PMID: 30683859
- PMCID: PMC6347618
- DOI: 10.1038/s41467-019-08390-z
The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma
Abstract
MicroRNA deregulation is a consistent feature of glioblastoma, yet the biological effect of each single gene is generally modest, and therapeutically negligible. Here we describe a module of microRNAs, constituted by miR-124, miR-128 and miR-137, which are co-expressed during neuronal differentiation and simultaneously lost in gliomagenesis. Each one of these miRs targets several transcriptional regulators, including the oncogenic chromatin repressors EZH2, BMI1 and LSD1, which are functionally interdependent and involved in glioblastoma recurrence after therapeutic chemoradiation. Synchronizing the expression of these three microRNAs in a gene therapy approach displays significant anticancer synergism, abrogates this epigenetic-mediated, multi-protein tumor survival mechanism and results in a 5-fold increase in survival when combined with chemotherapy in murine glioblastoma models. These transgenic microRNA clusters display intercellular propagation in vivo, via extracellular vesicles, extending their biological effect throughout the whole tumor. Our results support the rationale and feasibility of combinatorial microRNA strategies for anticancer therapies.
Conflict of interest statement
E.A.C. is currently an advisor to Advantagene Inc., Alcyone Biosciences, Insightec, Inc., Sigilon Therepeutics and DNAtrix Inc. and has equity interest in DNAtrix; he has also advised Oncorus, Merck, Tocagen, Ziopharm, Stemgen, NanoTx., Ziopharm Oncology, Cerebral Therapeutics, Genenta, Janssen, Karcinolysis, and Shanghai Biotech. The other authors declare no competing interests.
Figures
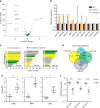

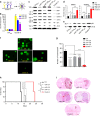

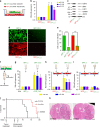
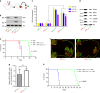
Comment in
-
Towards a microRNA-based Gene Therapy for Glioblastoma.Neurosurgery. 2019 Aug 1;85(2):E210-E211. doi: 10.1093/neuros/nyz166. Neurosurgery. 2019. PMID: 31304544 No abstract available.
References
-
- Chan JA, Krichevsky AM. Kosik KS.MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. - PubMed
-
- Ciafrè SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. - PubMed
-
- Nikaki A, Piperi C, Papavassiliou AG. Role of microRNAs in gliomagenesis: targeting miRNAs in glioblastoma multiforme therapy. Expert Opin. Investig. Drugs. 2012;21:1475–1488. - PubMed
-
- Ahir BK, Ozer H, Engelhard HH, Lakka SS. MicroRNAs in glioblastoma pathogenesis and therapy: a comprehensive review. Crit. Rev. Oncol. Hematol. 2017;120:22–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

