Evidence for detection of rat P2X4 receptor expressed on cells by generating monoclonal antibodies recognizing the native structure
- PMID: 30684150
- PMCID: PMC6439026
- DOI: 10.1007/s11302-019-09646-5
Evidence for detection of rat P2X4 receptor expressed on cells by generating monoclonal antibodies recognizing the native structure
Abstract
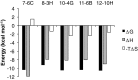
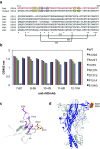
P2X purinergic receptors are ATP-driven ionic channels expressed as trimers and showing various functions. A subtype, the P2X4 receptor present on microglial cells is highly involved in neuropathic pain. In this study, in order to prepare antibodies recognizing the native structure of rat P2X4 (rP2X4) receptor, we immunized mice with rP2X4's head domain (rHD, Gln111-Val167), which possesses an intact structure stabilized by S-S bond formation (Igawa and Abe et al. FEBS Lett. 2015), as an antigen. We generated five monoclonal antibodies with the ability to recognize the native structure of its head domain, stabilized by S-S bond formation. Site-directed mutagenesis revealed that Asn127 and Asp131 of the rHD, in which combination of these amino acid residues is only conserved in P2X4 receptor among P2X family, were closely involved in the interaction between rHD and these antibodies. We also demonstrated the antibodies obtained here could detect rP2X4 receptor expressed in 1321N1 human astrocytoma cells.
Keywords: FSEC; Monoclonal antibody; Neuropathic pain; P2X4 receptor.
Conflict of interest statement
Conflicts of interest
Tatsuhiro Igawa declares that he has no conflict of interest.
Shuhei Kishikawa declares that he has no conflict of interest.
Yoshito Abe declares that he has no conflict of interest.
Tomohiro Yamashita declares that he has no conflict of interest.
Saki Nagai declares that she has no conflict of interest.
Mitsunori Shiroishi declares that he has no conflict of interest.
Chinatsu Shinozaki declares that he has no conflict of interest.
Hiroyuki Tanaka declares that he has no conflict of interest.
Hidetoshi Tozaki-Saitoh declares that he has no conflict of interest.
Makoto Tsuda declares that he has no conflict of interest.
Kazuhide Inoue declares that he has no conflict of interest.
Tadashi Ueda declares that he has no conflict of interest.
Ethical approval
All animal experiments were conducted according to the relevant national and international guidelines in the Act on Welfare and Management of Animals (Ministry of Environment of Japan) and the Regulation of Laboratory Animals (Kyushu University), and under the protocols approved by the Institutional Animal Care and Use Committee review panels at Kyushu University.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

