Next-generation sequencing identifies novel mitochondrial variants in pituitary adenomas
- PMID: 30684245
- PMCID: PMC6647476
- DOI: 10.1007/s40618-019-1005-6
Next-generation sequencing identifies novel mitochondrial variants in pituitary adenomas
Abstract
Purpose: Disrupted mitochondrial functions and genetic variants of mitochondrial DNA (mtDNA) have been observed in different human neoplasms. Next-generation sequencing (NGS) can be used to detect even low heteroplasmy-level mtDNA variants. We aimed to investigate the mitochondrial genome in pituitary adenomas by NGS.
Methods: We analysed 11 growth hormone producing and 33 non-functioning [22 gonadotroph and 11 hormone immunonegative] pituitary adenomas using VariantPro™ Mitochondrion Panel on Illumina MiSeq instrument. Revised Cambridge Reference Sequence (rCRS) of the mtDNA was used as reference. Heteroplasmy was determined using a 3% cutoff.
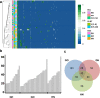
Results: 496 variants were identified in pituitary adenomas with overall low level of heteroplasmy (7.22%). On average, 35 variants were detected per sample. Samples harbouring the highest number of variants had the highest Ki-67 indices independently of histological subtypes. We identified eight variants (A11251G, T4216C, T16126C, C15452A, T14798C, A188G, G185A, and T16093C) with different prevalences among different histological groups. T16189C was found in 40% of non-recurrent adenomas, while it was not present in the recurrent ones. T14798C and T4216C were confirmed by Sanger sequencing in all 44 samples. 100% concordance was found between NGS and Sanger method.
Conclusions: NGS is a reliable method for investigating mitochondrial genome and heteroplasmy in pituitary adenomas. Out of the 496 detected variants, 414 have not been previously reported in pituitary adenoma. The high number of mtDNA variants may contribute to adenoma genesis, and some variants (i.e., T16189C) might associate with benign behaviour.
Keywords: Genetic variations; Genome; Mitochondria; Next-generation sequencing; Pathogenesis; Pituitary adenoma.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

