The cis-Regulatory Atlas of the Mouse Immune System
- PMID: 30686579
- PMCID: PMC6785993
- DOI: 10.1016/j.cell.2018.12.036
The cis-Regulatory Atlas of the Mouse Immune System
Abstract
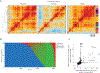
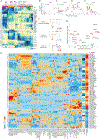
A complete chart of cis-regulatory elements and their dynamic activity is necessary to understand the transcriptional basis of differentiation and function of an organ system. We generated matched epigenome and transcriptome measurements in 86 primary cell types that span the mouse immune system and its differentiation cascades. This breadth of data enable variance components analysis that suggests that genes fall into two distinct classes, controlled by either enhancer- or promoter-driven logic, and multiple regression that connects genes to the enhancers that regulate them. Relating transcription factor (TF) expression to the genome-wide accessibility of their binding motifs classifies them as predominantly openers or closers of local chromatin accessibility, pinpointing specific cis-regulatory elements where binding of given TFs is likely functionally relevant, validated by chromatin immunoprecipitation sequencing (ChIP-seq). Overall, this cis-regulatory atlas provides a trove of information on transcriptional regulation through immune differentiation and a foundational scaffold to define key regulatory events throughout the immunological genome.
Keywords: ATAC-seq; Transcriptional regulation; chromatin; enhancer; epigenomics; immune cell differentiation; transcription factor.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures





References
-
- Anscombe FJ (1948). The Transformation of Poisson, Binomial and Negative-Binomial Data. Biometrika 35, 246–254.
-
- Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, et al. (2018). A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520. - PubMed
-
- Banerji J, Rusconi S, and Schaffner W (1981). Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308. - PubMed
-
- Benoist C, and Chambon P (1981). In vivo sequence requirements of the SV40 early promotor region. Nature 290, 304–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

