Genetic Screens Reveal FEN1 and APEX2 as BRCA2 Synthetic Lethal Targets
- PMID: 30686591
- PMCID: PMC6892393
- DOI: 10.1016/j.molcel.2018.12.008
Genetic Screens Reveal FEN1 and APEX2 as BRCA2 Synthetic Lethal Targets
Abstract
BRCA1 or BRCA2 inactivation drives breast and ovarian cancer but also creates vulnerability to poly(ADP-ribose) polymerase (PARP) inhibitors. To search for additional targets whose inhibition is synthetically lethal in BRCA2-deficient backgrounds, we screened two pairs of BRCA2 isogenic cell lines with DNA-repair-focused small hairpin RNA (shRNA) and CRISPR (clustered regularly interspaced short palindromic repeats)-based libraries. We found that BRCA2-deficient cells are selectively dependent on multiple pathways including base excision repair, ATR signaling, and splicing. We identified APEX2 and FEN1 as synthetic lethal genes with both BRCA1 and BRCA2 loss of function. BRCA2-deficient cells require the apurinic endonuclease activity and the PCNA-binding domain of Ape2 (APEX2), but not Ape1 (APEX1). Furthermore, BRCA2-deficient cells require the 5' flap endonuclease but not the 5'-3' exonuclease activity of Fen1, and chemically inhibiting Fen1 selectively targets BRCA-deficient cells. Finally, we developed a microhomology-mediated end-joining (MMEJ) reporter and showed that Fen1 participates in MMEJ, underscoring the importance of MMEJ as a collateral repair pathway in the context of homologous recombination (HR) deficiency.
Keywords: APEX2; ATR; BER; BRCA1; BRCA2; FEN1; MMEJ; NHEJ; PARP; synthetic lethality.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures
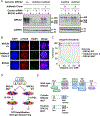



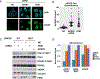
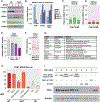

References
-
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, and Aguilera A (2014). BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

