Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases
- PMID: 30686769
- PMCID: PMC6398439
- DOI: 10.1016/j.ccell.2018.12.010
Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases
Abstract
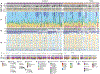
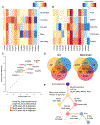
We integrated clinical, genomic, and transcriptomic data from 224 primaries and 95 metastases from 289 patients to characterize progression of pancreatic ductal adenocarcinoma (PDAC). Driver gene alterations and mutational and expression-based signatures were preserved, with truncations, inversions, and translocations most conserved. Cell cycle progression (CCP) increased with sequential inactivation of tumor suppressors, yet remained higher in metastases, perhaps driven by cell cycle regulatory gene variants. Half of the cases were hypoxic by expression markers, overlapping with molecular subtypes. Paired tumor heterogeneity showed cancer cell migration by Halstedian progression. Multiple PDACs arising synchronously and metachronously in the same pancreas were actually intra-parenchymal metastases, not independent primary tumors. Established clinical co-variates dominated survival analyses, although CCP and hypoxia may inform clinical practice.
Keywords: RNA sequencing; cell cycle progression; driver genes; hypoxia; metastases; mutational signatures; pancreatic ductal adenocarcinoma; whole-genome sequencing.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures






References
-
- ALMOGUERA C, SHIBATA D, FORRESTER K, MARTIN J, ARNHEIM N & PERUCHO M 1988. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell, 53, 549–54. - PubMed
-
- BAILEY P, CHANG DK, NONES K, JOHNS AL, PATCH AM, GINGRAS MC, MILLER DK, CHRIST AN, BRUXNER TJ, QUINN MC, et al. , 2016. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature, 531, 47–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

