IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections
- PMID: 30687247
- PMCID: PMC6338058
- DOI: 10.3389/fmicb.2018.03228
IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections
Abstract
Interferon-induced transmembrane proteins (IFITMs) are a family of small proteins that localize in the plasma and endolysosomal membranes. IFITMs not only inhibit viral entry into host cells by interrupting the membrane fusion between viral envelope and cellular membranes, but also reduce the production of infectious virions or infectivity of progeny virions. Not surprisingly, some viruses can evade the restriction of IFITMs and even hijack the antiviral proteins to facilitate their infectious entry into host cells or promote the assembly of virions, presumably by modulating membrane fusion. Similar to many other host defense genes that evolve under the selective pressure of microorganism infection, IFITM genes evolved in an accelerated speed in vertebrates and many single-nucleotide polymorphisms (SNPs) have been identified in the human population, some of which have been associated with severity and prognosis of viral infection (e.g., influenza A virus). Here, we review the function and potential impact of genetic variation for IFITM restriction of viral infections. Continuing research efforts are required to decipher the molecular mechanism underlying the complicated interaction among IFITMs and viruses in an effort to determine their pathobiological roles in the context of viral infections in vivo.
Keywords: IFITM; host susceptibility; interferon-induced transmembrane proteins; single nucleotide polymorphisms; viral infection.
Figures
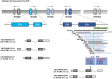
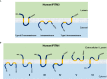
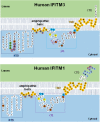
References
-
- Appourchaux R., Delpeuch M., Zhong L., Burlaud-Gaillard J., Tartour K., Savidis G., et al. (2018). Functional mapping of regions involved in the negative imprinting of virion particles infectivity and in target cell protection by the Interferon-Induced Transmembrane Protein 3 (IFITM3) against HIV-1. J. Virol. 10.1128/JVI.01716-18 [Epub ahead of print]. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

