Methodologies for Improving HDR Efficiency
- PMID: 30687381
- PMCID: PMC6338032
- DOI: 10.3389/fgene.2018.00691
Methodologies for Improving HDR Efficiency
Abstract
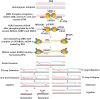
Clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) is a precise genome manipulating technology that can be programmed to induce double-strand break (DSB) in the genome wherever needed. After nuclease cleavage, DSBs can be repaired by non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathway. For producing targeted gene knock-in or other specific mutations, DSBs should be repaired by the HDR pathway. While NHEJ can cause various length insertions/deletion mutations (indels), which can lead the targeted gene to lose its function by shifting the open reading frame (ORF). Furthermore, HDR has low efficiency compared with the NHEJ pathway. In order to modify the gene precisely, numerous methods arose by inhibiting NHEJ or enhancing HDR, such as chemical modulation, synchronized expression, and overlapping homology arm. Here we focus on the efficiency and other considerations of these methodologies.
Keywords: CRISPR-Cas9; DSB; HDR; HDR enhancement; NHEJ; NHEJ inhibitors; cell arrest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

