A Comprehensive Review on the Interaction Between the Host GTPase Rab11 and Influenza A Virus
- PMID: 30687703
- PMCID: PMC6333742
- DOI: 10.3389/fcell.2018.00176
A Comprehensive Review on the Interaction Between the Host GTPase Rab11 and Influenza A Virus
Abstract
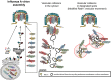
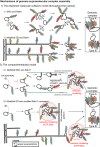
This year marks the 100th anniversary of one of the deadliest pandemic outbreaks, commonly referred as the Spanish Flu, that was caused by influenza A virus (IAV). Since then, IAV has been in governmental agendas worldwide, and a lot of effort has been put into understanding the pathogen's lifecycle, predict and mitigate the emergence of the strains that provoke yearly epidemics and pandemic events. Despite decades of research and seminal contributions there is still a lot to be investigated. In particular for this review, IAV lifecycle that takes place inside the host cell is not fully understood. Two steps that need clarification include genome transport to budding sites and genome assembly, the latter a complex process challenged by the nature of IAV genome that is divided into eight distinct parts. Assembly of such segmented genome is crucial to form fully infectious viral particles but is also critical for the emergence of viruses with pandemic potential that arise when avian and human IAV strains co-infect a host. The host GTPase Rab11 was separately implicated in both steps, and, interestingly these processes are beginning to emerge as being intimately related. Rab11 was initially proposed to be involved in the budding/release of IAV virions. It was subsequently shown to transport progeny genome, and later proposed to promote assembly of viral genome, but the underlying bridging mechanism the two is far from clear. For simplicity, this Rab11-centric review provides an initial separate account of Rab11 involvement in genome transport and in assembly. IAV genome assembly is a complicated molecular biology process, and therefore earned a dedicated section on how/if the viral genome forms a genomic supramolecular complex. Both topics present intricate challenges, outstanding questions, and unique controversies. At the end of the review, I will explore possible mechanisms intertwining IAV vRNP transport and genome assembly. Importantly, Rab11 has recently emerged as a key factor subverted by evolutionary unrelated viral families (Paramyxo, Bunya, and Orthomyxoviruses, among many others) and bacteria (Salmonella and Shigella) relevant to human health. This review provides a framework to identify common biological principles among the lifecycles of these pathogens.
Keywords: Rab11 GTPase; influenza A virus; influenza supramolecular genomic complex; viral assembly; viral inclusion.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources

