Identification of a pathogenic mutation in ATP2A1 via in silico analysis of exome data for cryptic aberrant splice sites
- PMID: 30688039
- PMCID: PMC6418371
- DOI: 10.1002/mgg3.552
Identification of a pathogenic mutation in ATP2A1 via in silico analysis of exome data for cryptic aberrant splice sites
Abstract
Background: Pathogenic mutations causing aberrant splicing are often difficult to detect. Standard variant analysis of next-generation sequence (NGS) data focuses on canonical splice sites. Noncanonical splice sites are more difficult to ascertain.
Methods: We developed a bioinformatics pipeline that screens existing NGS data for potentially aberrant novel essential splice sites (PANESS) and performed a pilot study on a family with a myotonic disorder. Further analyses were performed via qRT-PCR, immunoblotting, and immunohistochemistry. RNAi knockdown studies were performed in Drosophila to model the gene deficiency.
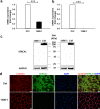
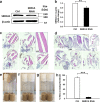
Results: The PANESS pipeline identified a homozygous ATP2A1 variant (NC_000016.9:g.28905928G>A; NM_004320.4:c.1287G>A:p.(Glu429=)) that was predicted to cause the omission of exon 11. Aberrant splicing of ATP2A1 was confirmed via qRT-PCR, and abnormal expression of the protein product sarcoplasmic/endoplasmic reticulum Ca++ ATPase 1 (SERCA1) was demonstrated in quadriceps femoris tissue from the proband. Ubiquitous knockdown of SERCA led to lethality in Drosophila, as did knockdown targeting differentiating or fusing myoblasts.
Conclusions: This study confirms the potential of novel in silico algorithms to detect cryptic mutations in existing NGS data; expands the phenotypic spectrum of ATP2A1 mutations beyond classic Brody myopathy; and suggests that genetic testing of ATP2A1 should be considered in patients with clinical myotonia.
Keywords: ATP2A1; Aberrant RNA splicing; Brody myopathy; cryptic variants.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Aarskog, N. K. , & Vedeler, C. A. (2000). Real‐time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot‐Marie‐Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Human Genetics, 107(5), 494–498. 10.1007/s004390000399 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

