Viral Vector-Based Delivery of CRISPR/Cas9 and Donor DNA for Homology-Directed Repair in an In Vitro Model for Canine Hemophilia B
- PMID: 30690229
- PMCID: PMC6356096
- DOI: 10.1016/j.omtn.2018.12.008
Viral Vector-Based Delivery of CRISPR/Cas9 and Donor DNA for Homology-Directed Repair in an In Vitro Model for Canine Hemophilia B
Abstract
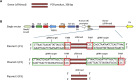
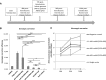
Gene therapy represents an attractive alternative to treat hemophilia B. Here we established three hepatocyte-derived cell lines based on Huh7, PLC/PRF/5, and Hep3B cells stably carrying a mutated canine FIX (cFIXmut) transgene containing a single point mutation in the catalytic domain. Based on these in vitro models resembling a commonly used canine large animal model, the tetracycline-controlled transcriptional activator (Tet-on)-inducible CRISPR/Cas9 system and an optimized donor were used to correct mutated cFIX gene through homology-directed repair (HDR). For efficient delivery of designer nuclease and donor DNA, we produced a high-capacity adenovirus vector type 5 (HCAdV5) containing the Tet-on-inducible cFIX-specific CRISPR/Cas9 system and a single-stranded adeno-associated virus type 2 vector (ssAAV2) containing the modified donor. Moreover, we designed a single HCAdV5 delivering all components for HDR. Our amplification-refractory mutation system based on qPCR analysis (ARMS-qPCR) revealed that the single vector application in Huh7-cFIXmut cells resulted in up to 5.52% HDR efficiencies, which was superior to the two-vector strategy. Furthermore the single vector also resulted in increased phenotypic correction efficiencies assayed by ELISA. We conclude that HDR in combination with viral vector delivery holds great promise for the correction of mutated FIX in disease models.
Keywords: CRISPR/Cas9; HCAdV5; gene therapy; hemophilia B; homology directed repair; single vector; ssAAV2.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Designer nuclease-mediated gene correction via homology-directed repair in an in vitro model of canine hemophilia B.J Gene Med. 2018 May;20(5):e3020. doi: 10.1002/jgm.3020. Epub 2018 May 3. J Gene Med. 2018. PMID: 29608237
-
Delivery of Cas9-guided ABE8e into stem cells using poly(l-lysine) polypeptides for correction of the hemophilia-associated FIX missense mutation.Biochem Biophys Res Commun. 2022 Nov 5;628:49-56. doi: 10.1016/j.bbrc.2022.08.076. Epub 2022 Aug 28. Biochem Biophys Res Commun. 2022. PMID: 36081278
-
CRISPR-Cas9-Mediated Genome Editing in Leishmania donovani.mBio. 2015 Jul 21;6(4):e00861. doi: 10.1128/mBio.00861-15. mBio. 2015. PMID: 26199327 Free PMC article.
-
What´s new in Gene Therapy of Hemophilia.Curr Gene Ther. 2018;18(2):107-114. doi: 10.2174/1566523218666180214162312. Curr Gene Ther. 2018. PMID: 29446741 Review.
-
CRISPR/Cas9-mediated correction of human genetic disease.Sci China Life Sci. 2017 May;60(5):447-457. doi: 10.1007/s11427-017-9032-4. Epub 2017 May 3. Sci China Life Sci. 2017. PMID: 28534256 Review.
Cited by
-
CRISPR medicine for blood disorders: Progress and challenges in delivery.Front Genome Ed. 2023 Jan 6;4:1037290. doi: 10.3389/fgeed.2022.1037290. eCollection 2022. Front Genome Ed. 2023. PMID: 36687779 Free PMC article. Review.
-
Exploring Advanced CRISPR Delivery Technologies for Therapeutic Genome Editing.Small Sci. 2024 Jul 25;4(10):2400192. doi: 10.1002/smsc.202400192. eCollection 2024 Oct. Small Sci. 2024. PMID: 40212235 Free PMC article.
-
Adenovirus vectors in hematopoietic stem cell genome editing.FEBS Lett. 2019 Dec;593(24):3623-3648. doi: 10.1002/1873-3468.13668. Epub 2019 Nov 20. FEBS Lett. 2019. PMID: 31705806 Free PMC article. Review.
-
Gene Therapy Approaches for the Treatment of Hemophilia B.Int J Mol Sci. 2023 Jun 28;24(13):10766. doi: 10.3390/ijms241310766. Int J Mol Sci. 2023. PMID: 37445943 Free PMC article. Review.
-
The use of adenoviral vectors in gene therapy and vaccine approaches.Genet Mol Biol. 2022 Oct 7;45(3 Suppl 1):e20220079. doi: 10.1590/1678-4685-GMB-2022-0079. eCollection 2022. Genet Mol Biol. 2022. PMID: 36206378 Free PMC article.
References
-
- Nathwani A.C., Tuddenham E.G. Epidemiology of coagulation disorders. Baillieres Clin. Haematol. 1992;5:383–439. - PubMed
-
- Kay M.A., Rothenberg S., Landen C.N., Bellinger D.A., Leland F., Toman C., Finegold M., Thompson A.R., Read M.S., Brinkhous K.M. In vivo gene therapy of hemophilia B: sustained partial correction in factor IX-deficient dogs. Science. 1993;262:117–119. - PubMed
-
- Kay M.A., Manno C.S., Ragni M.V., Larson P.J., Couto L.B., McClelland A., Glader B., Chew A.J., Tai S.J., Herzog R.W. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 2000;24:257–261. - PubMed
LinkOut - more resources
Full Text Sources

